| Full name: complexin 3 | Alias Symbol: CPX-III | ||
| Type: protein-coding gene | Cytoband: 15q24.1 | ||
| Entrez ID: 594855 | HGNC ID: HGNC:27652 | Ensembl Gene: ENSG00000213578 | OMIM ID: 609585 |
| Drug and gene relationship at DGIdb | |||
Expression of CPLX3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CPLX3 | 594855 | 219775_s_at | 0.0063 | 0.9819 | |
| GSE20347 | CPLX3 | 594855 | 219775_s_at | 0.0601 | 0.3674 | |
| GSE23400 | CPLX3 | 594855 | 219775_s_at | -0.0219 | 0.4639 | |
| GSE26886 | CPLX3 | 594855 | 219775_s_at | 0.3867 | 0.0474 | |
| GSE29001 | CPLX3 | 594855 | 219775_s_at | -0.0512 | 0.8276 | |
| GSE38129 | CPLX3 | 594855 | 219775_s_at | -0.0344 | 0.6426 | |
| GSE45670 | CPLX3 | 594855 | 222927_s_at | -0.0379 | 0.8023 | |
| GSE63941 | CPLX3 | 594855 | 222927_s_at | 0.1338 | 0.4188 | |
| GSE77861 | CPLX3 | 594855 | 219775_s_at | -0.0389 | 0.8110 | |
| TCGA | CPLX3 | 594855 | RNAseq | -0.1983 | 0.8207 |
Upregulated datasets: 0; Downregulated datasets: 0.
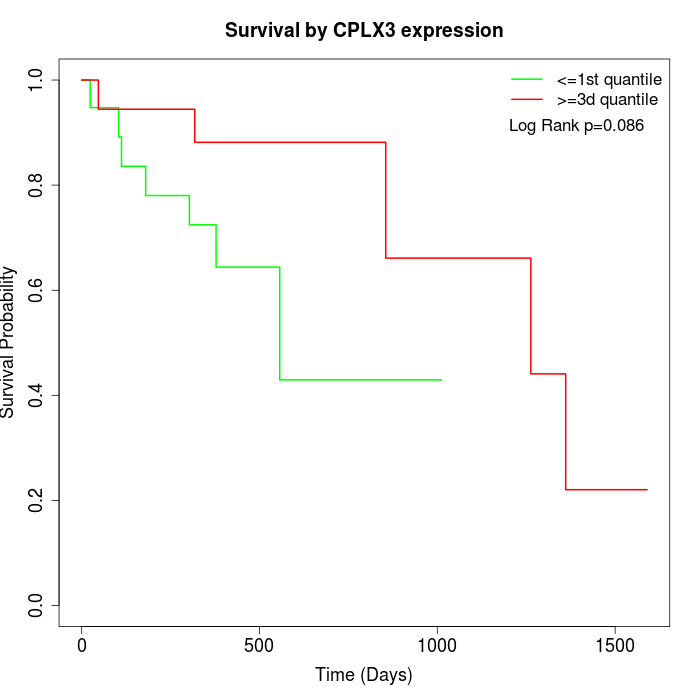
Survival by CPLX3 expression:
Note: Click image to view full size file.
Copy number change of CPLX3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CPLX3 | 594855 | 8 | 1 | 21 | |
| GSE20123 | CPLX3 | 594855 | 8 | 1 | 21 | |
| GSE43470 | CPLX3 | 594855 | 4 | 6 | 33 | |
| GSE46452 | CPLX3 | 594855 | 3 | 7 | 49 | |
| GSE47630 | CPLX3 | 594855 | 8 | 10 | 22 | |
| GSE54993 | CPLX3 | 594855 | 5 | 6 | 59 | |
| GSE54994 | CPLX3 | 594855 | 5 | 7 | 41 | |
| GSE60625 | CPLX3 | 594855 | 4 | 0 | 7 | |
| GSE74703 | CPLX3 | 594855 | 4 | 3 | 29 | |
| GSE74704 | CPLX3 | 594855 | 4 | 1 | 15 | |
| TCGA | CPLX3 | 594855 | 14 | 15 | 67 |
Total number of gains: 67; Total number of losses: 57; Total Number of normals: 364.
Somatic mutations of CPLX3:
Generating mutation plots.
Highly correlated genes for CPLX3:
Showing top 20/537 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CPLX3 | DUSP15 | 0.713559 | 3 | 0 | 3 |
| CPLX3 | CDRT15 | 0.683131 | 3 | 0 | 3 |
| CPLX3 | IQCC | 0.680545 | 4 | 0 | 3 |
| CPLX3 | RPRML | 0.673076 | 3 | 0 | 3 |
| CPLX3 | REG3A | 0.672548 | 3 | 0 | 3 |
| CPLX3 | ENTPD2 | 0.670133 | 5 | 0 | 4 |
| CPLX3 | GRIN1 | 0.665969 | 5 | 0 | 4 |
| CPLX3 | ARVCF | 0.663868 | 5 | 0 | 4 |
| CPLX3 | LCN1 | 0.661107 | 3 | 0 | 3 |
| CPLX3 | ARC | 0.659171 | 4 | 0 | 3 |
| CPLX3 | BRSK1 | 0.65729 | 3 | 0 | 3 |
| CPLX3 | FUT7 | 0.654952 | 5 | 0 | 4 |
| CPLX3 | CNGB1 | 0.653539 | 6 | 0 | 5 |
| CPLX3 | CEMP1 | 0.652755 | 6 | 0 | 5 |
| CPLX3 | TBX10 | 0.65193 | 5 | 0 | 5 |
| CPLX3 | C9orf131 | 0.651616 | 3 | 0 | 3 |
| CPLX3 | DNAI1 | 0.651178 | 3 | 0 | 3 |
| CPLX3 | MAPRE3 | 0.649691 | 6 | 0 | 5 |
| CPLX3 | CACTIN | 0.649406 | 3 | 0 | 3 |
| CPLX3 | NEUROD6 | 0.64834 | 4 | 0 | 4 |
For details and further investigation, click here