| Full name: chondroitin sulfate N-acetylgalactosaminyltransferase 1 | Alias Symbol: CSGalNAcT-1|FLJ11264|ChGn | ||
| Type: protein-coding gene | Cytoband: 8p21.3 | ||
| Entrez ID: 55790 | HGNC ID: HGNC:24290 | Ensembl Gene: ENSG00000147408 | OMIM ID: 616615 |
| Drug and gene relationship at DGIdb | |||
Expression of CSGALNACT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CSGALNACT1 | 55790 | 219049_at | -1.2477 | 0.1947 | |
| GSE20347 | CSGALNACT1 | 55790 | 219049_at | 0.2198 | 0.4765 | |
| GSE23400 | CSGALNACT1 | 55790 | 219049_at | -0.0583 | 0.7377 | |
| GSE26886 | CSGALNACT1 | 55790 | 219049_at | 1.7051 | 0.0105 | |
| GSE29001 | CSGALNACT1 | 55790 | 219049_at | 0.5234 | 0.3761 | |
| GSE38129 | CSGALNACT1 | 55790 | 219049_at | -0.3827 | 0.3006 | |
| GSE45670 | CSGALNACT1 | 55790 | 219049_at | -1.7618 | 0.0009 | |
| GSE53622 | CSGALNACT1 | 55790 | 104622 | -0.9824 | 0.0000 | |
| GSE53624 | CSGALNACT1 | 55790 | 104622 | -0.7925 | 0.0000 | |
| GSE63941 | CSGALNACT1 | 55790 | 219049_at | -5.2022 | 0.0001 | |
| GSE77861 | CSGALNACT1 | 55790 | 219049_at | 1.6052 | 0.0206 | |
| GSE97050 | CSGALNACT1 | 55790 | A_33_P3366540 | 0.1420 | 0.6190 | |
| SRP007169 | CSGALNACT1 | 55790 | RNAseq | 1.5059 | 0.0056 | |
| SRP008496 | CSGALNACT1 | 55790 | RNAseq | 2.7778 | 0.0000 | |
| SRP064894 | CSGALNACT1 | 55790 | RNAseq | -0.4229 | 0.1605 | |
| SRP133303 | CSGALNACT1 | 55790 | RNAseq | -1.0033 | 0.0000 | |
| SRP159526 | CSGALNACT1 | 55790 | RNAseq | -0.6344 | 0.0352 | |
| SRP193095 | CSGALNACT1 | 55790 | RNAseq | 1.0752 | 0.0000 | |
| SRP219564 | CSGALNACT1 | 55790 | RNAseq | -0.7646 | 0.1548 | |
| TCGA | CSGALNACT1 | 55790 | RNAseq | -0.5108 | 0.0000 |
Upregulated datasets: 5; Downregulated datasets: 3.
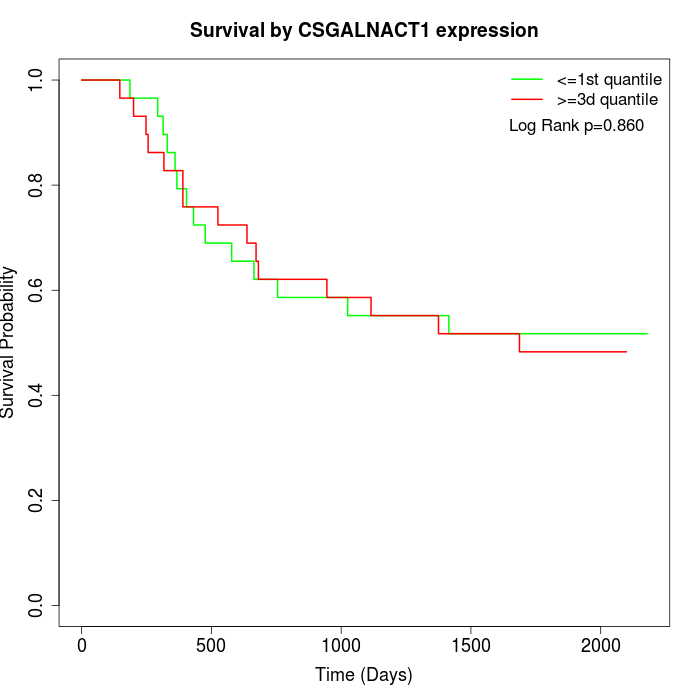
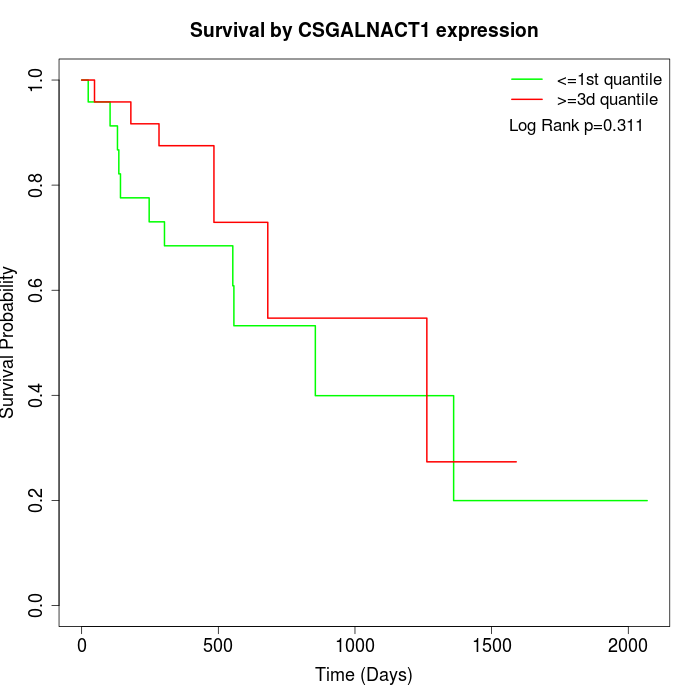
Survival by CSGALNACT1 expression:
Note: Click image to view full size file.
Copy number change of CSGALNACT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CSGALNACT1 | 55790 | 4 | 10 | 16 | |
| GSE20123 | CSGALNACT1 | 55790 | 4 | 10 | 16 | |
| GSE43470 | CSGALNACT1 | 55790 | 4 | 8 | 31 | |
| GSE46452 | CSGALNACT1 | 55790 | 14 | 13 | 32 | |
| GSE47630 | CSGALNACT1 | 55790 | 10 | 8 | 22 | |
| GSE54993 | CSGALNACT1 | 55790 | 2 | 14 | 54 | |
| GSE54994 | CSGALNACT1 | 55790 | 8 | 18 | 27 | |
| GSE60625 | CSGALNACT1 | 55790 | 3 | 0 | 8 | |
| GSE74703 | CSGALNACT1 | 55790 | 4 | 7 | 25 | |
| GSE74704 | CSGALNACT1 | 55790 | 3 | 6 | 11 | |
| TCGA | CSGALNACT1 | 55790 | 14 | 43 | 39 |
Total number of gains: 70; Total number of losses: 137; Total Number of normals: 281.
Somatic mutations of CSGALNACT1:
Generating mutation plots.
Highly correlated genes for CSGALNACT1:
Showing top 20/799 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CSGALNACT1 | CNRIP1 | 0.779969 | 6 | 0 | 6 |
| CSGALNACT1 | DNM3OS | 0.755436 | 3 | 0 | 3 |
| CSGALNACT1 | EBF1 | 0.751787 | 7 | 0 | 7 |
| CSGALNACT1 | LINC01082 | 0.736182 | 5 | 0 | 5 |
| CSGALNACT1 | NEDD9 | 0.729882 | 5 | 0 | 5 |
| CSGALNACT1 | TNFSF4 | 0.714773 | 3 | 0 | 3 |
| CSGALNACT1 | IL13RA2 | 0.712967 | 3 | 0 | 3 |
| CSGALNACT1 | GNG2 | 0.711139 | 5 | 0 | 5 |
| CSGALNACT1 | RHOJ | 0.71054 | 4 | 0 | 4 |
| CSGALNACT1 | CCDC3 | 0.707873 | 5 | 0 | 5 |
| CSGALNACT1 | LINC01279 | 0.707852 | 4 | 0 | 4 |
| CSGALNACT1 | FZD4 | 0.705362 | 7 | 0 | 7 |
| CSGALNACT1 | ZEB2 | 0.701648 | 10 | 0 | 8 |
| CSGALNACT1 | TK2 | 0.69869 | 3 | 0 | 3 |
| CSGALNACT1 | NOL9 | 0.695666 | 3 | 0 | 3 |
| CSGALNACT1 | SGCD | 0.695534 | 6 | 0 | 6 |
| CSGALNACT1 | SLITRK6 | 0.695474 | 4 | 0 | 4 |
| CSGALNACT1 | ZBED1 | 0.693787 | 3 | 0 | 3 |
| CSGALNACT1 | MICU3 | 0.692555 | 5 | 0 | 5 |
| CSGALNACT1 | HNRNPUL2 | 0.690888 | 3 | 0 | 3 |
For details and further investigation, click here