| Full name: DDB1 and CUL4 associated factor 15 | Alias Symbol: MGC99481 | ||
| Type: protein-coding gene | Cytoband: 19p13.12 | ||
| Entrez ID: 90379 | HGNC ID: HGNC:25095 | Ensembl Gene: ENSG00000132017 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of DCAF15:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DCAF15 | 90379 | 91952_at | 0.3762 | 0.2028 | |
| GSE20347 | DCAF15 | 90379 | 91952_at | 0.2835 | 0.0193 | |
| GSE23400 | DCAF15 | 90379 | 91952_at | 0.4132 | 0.0000 | |
| GSE26886 | DCAF15 | 90379 | 222012_at | 0.2504 | 0.0921 | |
| GSE29001 | DCAF15 | 90379 | 91952_at | 0.3842 | 0.1775 | |
| GSE38129 | DCAF15 | 90379 | 91952_at | 0.3417 | 0.0279 | |
| GSE45670 | DCAF15 | 90379 | 91952_at | 0.1199 | 0.1003 | |
| GSE53622 | DCAF15 | 90379 | 3017 | 0.1293 | 0.2309 | |
| GSE53624 | DCAF15 | 90379 | 3017 | 0.4160 | 0.0000 | |
| GSE63941 | DCAF15 | 90379 | 91952_at | 1.0655 | 0.0008 | |
| GSE77861 | DCAF15 | 90379 | 222012_at | -0.0543 | 0.6952 | |
| GSE97050 | DCAF15 | 90379 | A_23_P50399 | -0.2514 | 0.5552 | |
| SRP007169 | DCAF15 | 90379 | RNAseq | 0.1188 | 0.7950 | |
| SRP008496 | DCAF15 | 90379 | RNAseq | -0.5301 | 0.1482 | |
| SRP064894 | DCAF15 | 90379 | RNAseq | 0.7752 | 0.0015 | |
| SRP133303 | DCAF15 | 90379 | RNAseq | 0.0589 | 0.7525 | |
| SRP159526 | DCAF15 | 90379 | RNAseq | 0.0929 | 0.7872 | |
| SRP193095 | DCAF15 | 90379 | RNAseq | 0.3883 | 0.0000 | |
| SRP219564 | DCAF15 | 90379 | RNAseq | 0.5502 | 0.1752 | |
| TCGA | DCAF15 | 90379 | RNAseq | 0.3491 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
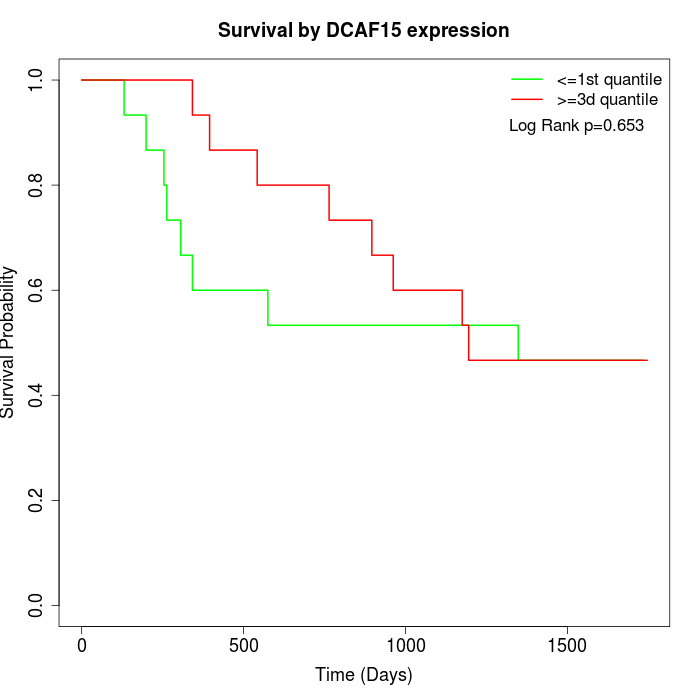
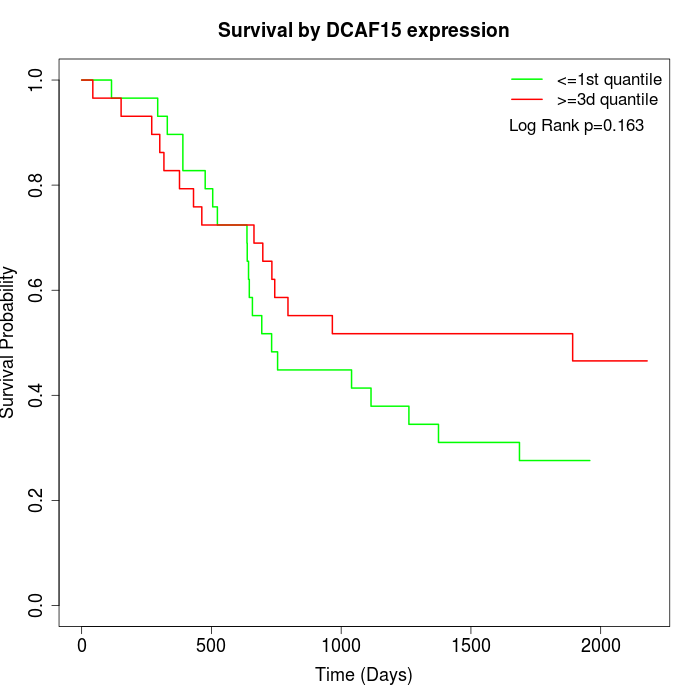
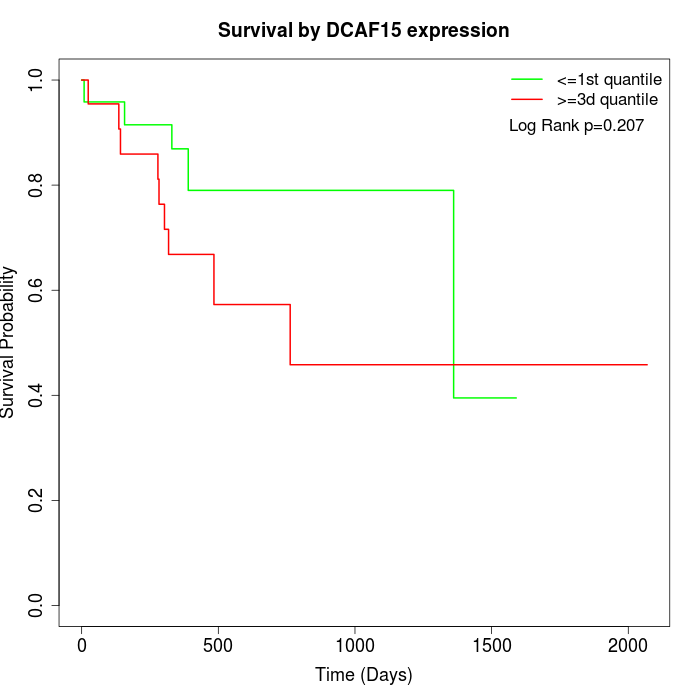
Survival by DCAF15 expression:
Note: Click image to view full size file.
Copy number change of DCAF15:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DCAF15 | 90379 | 4 | 2 | 24 | |
| GSE20123 | DCAF15 | 90379 | 3 | 1 | 26 | |
| GSE43470 | DCAF15 | 90379 | 2 | 6 | 35 | |
| GSE46452 | DCAF15 | 90379 | 47 | 1 | 11 | |
| GSE47630 | DCAF15 | 90379 | 4 | 7 | 29 | |
| GSE54993 | DCAF15 | 90379 | 15 | 4 | 51 | |
| GSE54994 | DCAF15 | 90379 | 6 | 13 | 34 | |
| GSE60625 | DCAF15 | 90379 | 9 | 0 | 2 | |
| GSE74703 | DCAF15 | 90379 | 2 | 4 | 30 | |
| GSE74704 | DCAF15 | 90379 | 0 | 1 | 19 | |
| TCGA | DCAF15 | 90379 | 22 | 11 | 63 |
Total number of gains: 114; Total number of losses: 50; Total Number of normals: 324.
Somatic mutations of DCAF15:
Generating mutation plots.
Highly correlated genes for DCAF15:
Showing top 20/466 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DCAF15 | CDH4 | 0.799554 | 3 | 0 | 3 |
| DCAF15 | JSRP1 | 0.770436 | 3 | 0 | 3 |
| DCAF15 | HSD17B14 | 0.754214 | 3 | 0 | 3 |
| DCAF15 | SNORA71A | 0.743615 | 3 | 0 | 3 |
| DCAF15 | TXNDC2 | 0.723047 | 3 | 0 | 3 |
| DCAF15 | MGAT5B | 0.718896 | 4 | 0 | 4 |
| DCAF15 | FGF18 | 0.718322 | 3 | 0 | 3 |
| DCAF15 | UFSP1 | 0.717461 | 3 | 0 | 3 |
| DCAF15 | RNF151 | 0.715411 | 3 | 0 | 3 |
| DCAF15 | NKAIN4 | 0.713978 | 4 | 0 | 3 |
| DCAF15 | RFT1 | 0.708271 | 3 | 0 | 3 |
| DCAF15 | FAM178B | 0.705518 | 3 | 0 | 3 |
| DCAF15 | CLTCL1 | 0.703394 | 3 | 0 | 3 |
| DCAF15 | RAD51D | 0.702313 | 3 | 0 | 3 |
| DCAF15 | REG3A | 0.700251 | 4 | 0 | 3 |
| DCAF15 | MEGF10 | 0.699106 | 3 | 0 | 3 |
| DCAF15 | C8orf74 | 0.698509 | 3 | 0 | 3 |
| DCAF15 | MAMSTR | 0.693457 | 3 | 0 | 3 |
| DCAF15 | FAM171A2 | 0.688662 | 4 | 0 | 3 |
| DCAF15 | SFTPD | 0.683082 | 3 | 0 | 3 |
For details and further investigation, click here