| Full name: early growth response 4 | Alias Symbol: NGFI-C|PAT133 | ||
| Type: protein-coding gene | Cytoband: 2p13.2 | ||
| Entrez ID: 1961 | HGNC ID: HGNC:3241 | Ensembl Gene: ENSG00000135625 | OMIM ID: 128992 |
| Drug and gene relationship at DGIdb | |||
Expression of EGR4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EGR4 | 1961 | 207767_s_at | 0.1845 | 0.4816 | |
| GSE20347 | EGR4 | 1961 | 207767_s_at | 0.0703 | 0.3871 | |
| GSE23400 | EGR4 | 1961 | 207767_s_at | -0.0489 | 0.3250 | |
| GSE26886 | EGR4 | 1961 | 231575_at | 0.3118 | 0.0512 | |
| GSE29001 | EGR4 | 1961 | 207767_s_at | -0.2267 | 0.2245 | |
| GSE38129 | EGR4 | 1961 | 207767_s_at | -0.0227 | 0.8016 | |
| GSE45670 | EGR4 | 1961 | 231575_at | 0.0025 | 0.9860 | |
| GSE63941 | EGR4 | 1961 | 207767_s_at | 0.4775 | 0.1915 | |
| GSE77861 | EGR4 | 1961 | 231575_at | -0.0616 | 0.5519 | |
| GSE97050 | EGR4 | 1961 | A_23_P380318 | 0.0608 | 0.7808 | |
| TCGA | EGR4 | 1961 | RNAseq | 0.8256 | 0.1363 |
Upregulated datasets: 0; Downregulated datasets: 0.
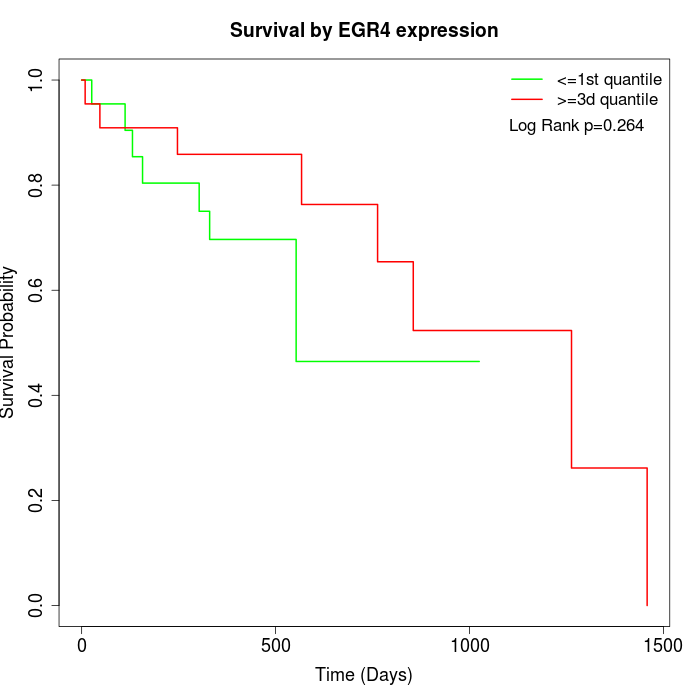
Survival by EGR4 expression:
Note: Click image to view full size file.
Copy number change of EGR4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EGR4 | 1961 | 11 | 1 | 18 | |
| GSE20123 | EGR4 | 1961 | 11 | 1 | 18 | |
| GSE43470 | EGR4 | 1961 | 4 | 0 | 39 | |
| GSE46452 | EGR4 | 1961 | 2 | 3 | 54 | |
| GSE47630 | EGR4 | 1961 | 7 | 0 | 33 | |
| GSE54993 | EGR4 | 1961 | 0 | 6 | 64 | |
| GSE54994 | EGR4 | 1961 | 11 | 0 | 42 | |
| GSE60625 | EGR4 | 1961 | 0 | 3 | 8 | |
| GSE74703 | EGR4 | 1961 | 4 | 0 | 32 | |
| GSE74704 | EGR4 | 1961 | 9 | 0 | 11 | |
| TCGA | EGR4 | 1961 | 38 | 2 | 56 |
Total number of gains: 97; Total number of losses: 16; Total Number of normals: 375.
Somatic mutations of EGR4:
Generating mutation plots.
Highly correlated genes for EGR4:
Showing top 20/1018 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EGR4 | PEX11G | 0.857599 | 3 | 0 | 3 |
| EGR4 | PPP1R13B | 0.794054 | 3 | 0 | 3 |
| EGR4 | PGC | 0.781286 | 4 | 0 | 4 |
| EGR4 | MYO15B | 0.757406 | 3 | 0 | 3 |
| EGR4 | STAC3 | 0.746836 | 3 | 0 | 3 |
| EGR4 | NXNL1 | 0.746427 | 3 | 0 | 3 |
| EGR4 | ZNF524 | 0.728579 | 3 | 0 | 3 |
| EGR4 | FOLR3 | 0.72493 | 3 | 0 | 3 |
| EGR4 | RHBG | 0.724768 | 3 | 0 | 3 |
| EGR4 | TMEM102 | 0.724642 | 3 | 0 | 3 |
| EGR4 | PROC | 0.721375 | 4 | 0 | 4 |
| EGR4 | TTLL9 | 0.72133 | 3 | 0 | 3 |
| EGR4 | OVGP1 | 0.713668 | 4 | 0 | 4 |
| EGR4 | ISLR2 | 0.712175 | 4 | 0 | 4 |
| EGR4 | GJD4 | 0.710533 | 4 | 0 | 3 |
| EGR4 | ABCC6 | 0.702562 | 4 | 0 | 3 |
| EGR4 | OR2F2 | 0.701708 | 3 | 0 | 3 |
| EGR4 | CNGB3 | 0.695159 | 6 | 0 | 5 |
| EGR4 | JSRP1 | 0.692362 | 4 | 0 | 3 |
| EGR4 | ZNF385A | 0.691308 | 3 | 0 | 3 |
For details and further investigation, click here