| Full name: progastricsin | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 6p21.1 | ||
| Entrez ID: 5225 | HGNC ID: HGNC:8890 | Ensembl Gene: ENSG00000096088 | OMIM ID: 169740 |
| Drug and gene relationship at DGIdb | |||
Expression of PGC:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PGC | 5225 | 205261_at | 0.1132 | 0.7379 | |
| GSE20347 | PGC | 5225 | 205261_at | -0.2355 | 0.2816 | |
| GSE23400 | PGC | 5225 | 205261_at | -0.4797 | 0.0093 | |
| GSE26886 | PGC | 5225 | 205261_at | -0.4858 | 0.0806 | |
| GSE29001 | PGC | 5225 | 205261_at | -0.2696 | 0.2133 | |
| GSE38129 | PGC | 5225 | 205261_at | -1.0130 | 0.0545 | |
| GSE45670 | PGC | 5225 | 205261_at | 0.1909 | 0.0451 | |
| GSE53622 | PGC | 5225 | 101199 | -0.4454 | 0.0603 | |
| GSE53624 | PGC | 5225 | 101199 | -1.0308 | 0.0000 | |
| GSE63941 | PGC | 5225 | 205261_at | -0.0517 | 0.7710 | |
| GSE77861 | PGC | 5225 | 205261_at | -0.1846 | 0.1970 | |
| GSE97050 | PGC | 5225 | A_33_P3370714 | -0.1689 | 0.5613 | |
| TCGA | PGC | 5225 | RNAseq | -5.0930 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 2.
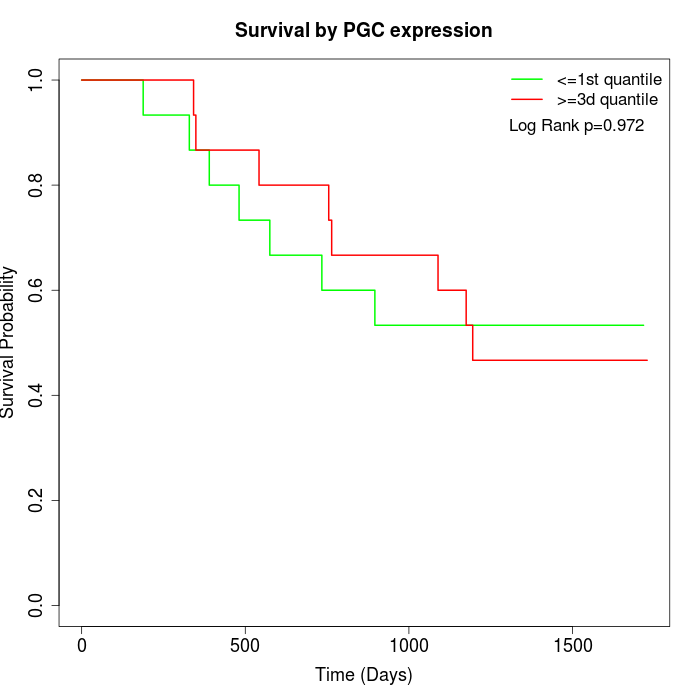
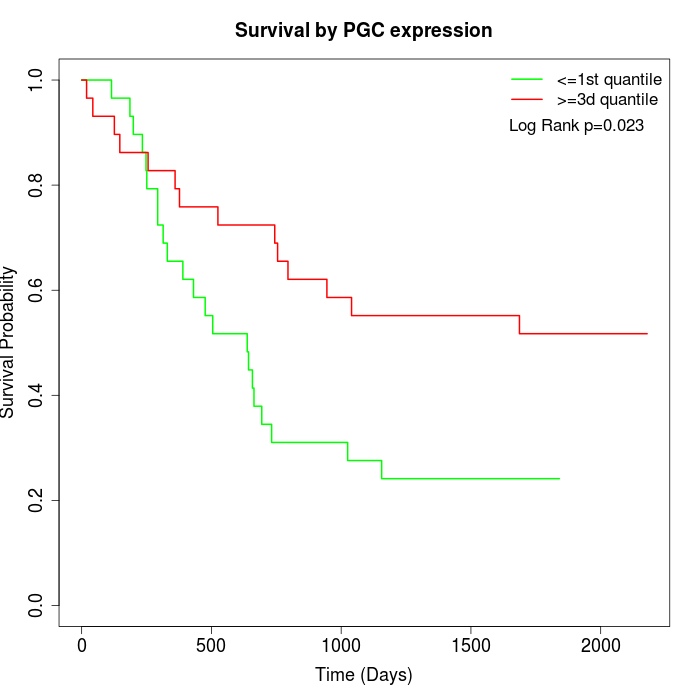
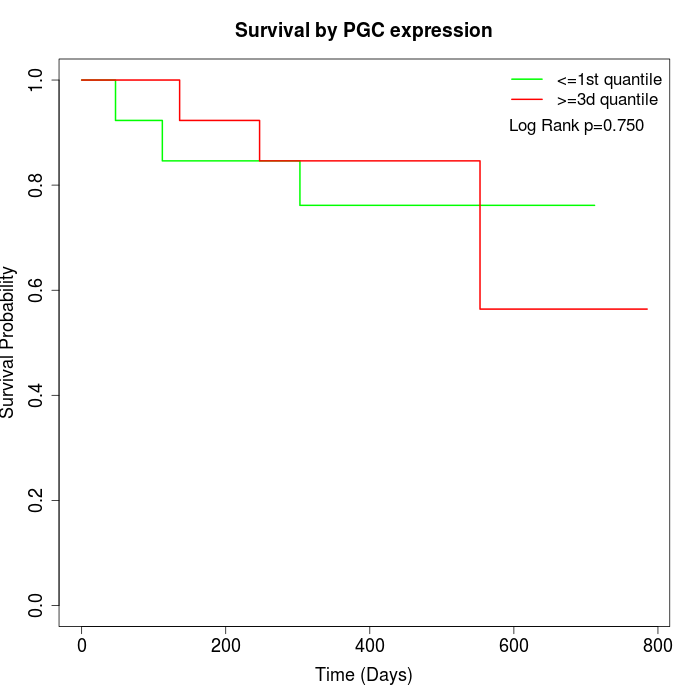
Survival by PGC expression:
Note: Click image to view full size file.
Copy number change of PGC:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PGC | 5225 | 6 | 1 | 23 | |
| GSE20123 | PGC | 5225 | 6 | 1 | 23 | |
| GSE43470 | PGC | 5225 | 6 | 0 | 37 | |
| GSE46452 | PGC | 5225 | 2 | 9 | 48 | |
| GSE47630 | PGC | 5225 | 8 | 4 | 28 | |
| GSE54993 | PGC | 5225 | 3 | 2 | 65 | |
| GSE54994 | PGC | 5225 | 11 | 4 | 38 | |
| GSE60625 | PGC | 5225 | 0 | 1 | 10 | |
| GSE74703 | PGC | 5225 | 6 | 0 | 30 | |
| GSE74704 | PGC | 5225 | 3 | 1 | 16 | |
| TCGA | PGC | 5225 | 21 | 13 | 62 |
Total number of gains: 72; Total number of losses: 36; Total Number of normals: 380.
Somatic mutations of PGC:
Generating mutation plots.
Highly correlated genes for PGC:
Showing top 20/416 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PGC | PHGR1 | 0.859643 | 4 | 0 | 4 |
| PGC | CPAMD8 | 0.842015 | 3 | 0 | 3 |
| PGC | FBLN7 | 0.829518 | 3 | 0 | 3 |
| PGC | KCNQ4 | 0.821099 | 3 | 0 | 3 |
| PGC | SIGLEC8 | 0.817535 | 3 | 0 | 3 |
| PGC | LIM2 | 0.815348 | 3 | 0 | 3 |
| PGC | CDX2 | 0.809062 | 3 | 0 | 3 |
| PGC | RNF123 | 0.808942 | 4 | 0 | 4 |
| PGC | TRIM10 | 0.808453 | 3 | 0 | 3 |
| PGC | CYP2D6 | 0.80647 | 4 | 0 | 4 |
| PGC | CHIT1 | 0.801402 | 3 | 0 | 3 |
| PGC | TULP1 | 0.796848 | 3 | 0 | 3 |
| PGC | LRCH4 | 0.795012 | 3 | 0 | 3 |
| PGC | MASP2 | 0.794995 | 3 | 0 | 3 |
| PGC | SLC18A3 | 0.793224 | 3 | 0 | 3 |
| PGC | ARVCF | 0.791054 | 3 | 0 | 3 |
| PGC | JAKMIP1 | 0.789836 | 3 | 0 | 3 |
| PGC | KLK2 | 0.789388 | 3 | 0 | 3 |
| PGC | NPY2R | 0.784527 | 3 | 0 | 3 |
| PGC | RCOR2 | 0.783025 | 3 | 0 | 3 |
For details and further investigation, click here