| Full name: euchromatic histone lysine methyltransferase 2 | Alias Symbol: G9A|Em:AF134726.3|NG36/G9a|KMT1C | ||
| Type: protein-coding gene | Cytoband: 6p21.33 | ||
| Entrez ID: 10919 | HGNC ID: HGNC:14129 | Ensembl Gene: ENSG00000204371 | OMIM ID: 604599 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
EHMT2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04211 | Longevity regulating pathway - mammal |
Expression of EHMT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EHMT2 | 10919 | 202326_at | 0.8430 | 0.1726 | |
| GSE20347 | EHMT2 | 10919 | 202326_at | 0.4303 | 0.0057 | |
| GSE23400 | EHMT2 | 10919 | 202326_at | 0.3819 | 0.0000 | |
| GSE26886 | EHMT2 | 10919 | 202326_at | 0.7520 | 0.0001 | |
| GSE29001 | EHMT2 | 10919 | 207484_s_at | -0.0927 | 0.5609 | |
| GSE38129 | EHMT2 | 10919 | 202326_at | 0.5347 | 0.0000 | |
| GSE45670 | EHMT2 | 10919 | 207484_s_at | -0.1250 | 0.2039 | |
| GSE53622 | EHMT2 | 10919 | 31281 | 0.3130 | 0.0000 | |
| GSE53624 | EHMT2 | 10919 | 31281 | 0.2291 | 0.0000 | |
| GSE63941 | EHMT2 | 10919 | 202326_at | 1.2501 | 0.0116 | |
| GSE77861 | EHMT2 | 10919 | 207484_s_at | -0.0278 | 0.8597 | |
| GSE97050 | EHMT2 | 10919 | A_23_P214638 | 0.1792 | 0.5291 | |
| SRP007169 | EHMT2 | 10919 | RNAseq | 0.3044 | 0.4292 | |
| SRP008496 | EHMT2 | 10919 | RNAseq | 0.5042 | 0.0562 | |
| SRP064894 | EHMT2 | 10919 | RNAseq | 0.5782 | 0.0056 | |
| SRP133303 | EHMT2 | 10919 | RNAseq | -0.1498 | 0.2888 | |
| SRP159526 | EHMT2 | 10919 | RNAseq | 0.5954 | 0.1146 | |
| SRP193095 | EHMT2 | 10919 | RNAseq | 0.3288 | 0.0008 | |
| SRP219564 | EHMT2 | 10919 | RNAseq | 0.4138 | 0.1616 | |
| TCGA | EHMT2 | 10919 | RNAseq | 0.1553 | 0.0012 |
Upregulated datasets: 1; Downregulated datasets: 0.
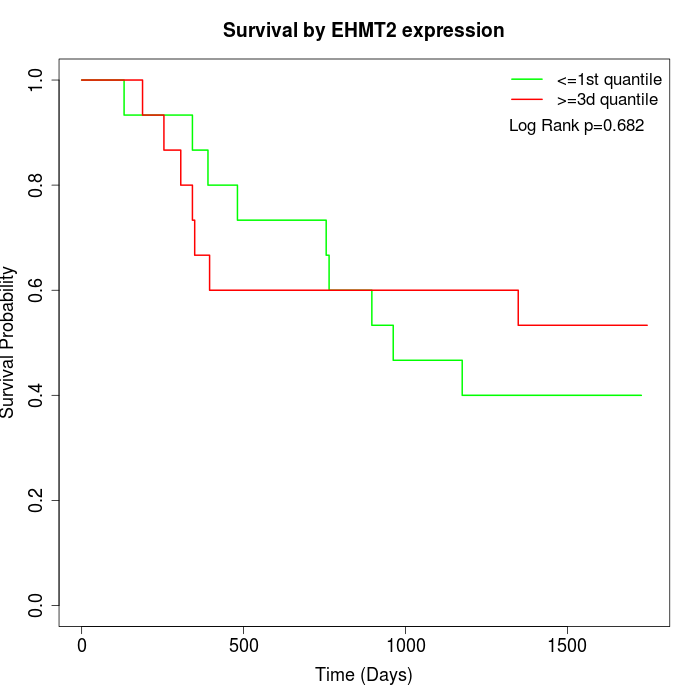
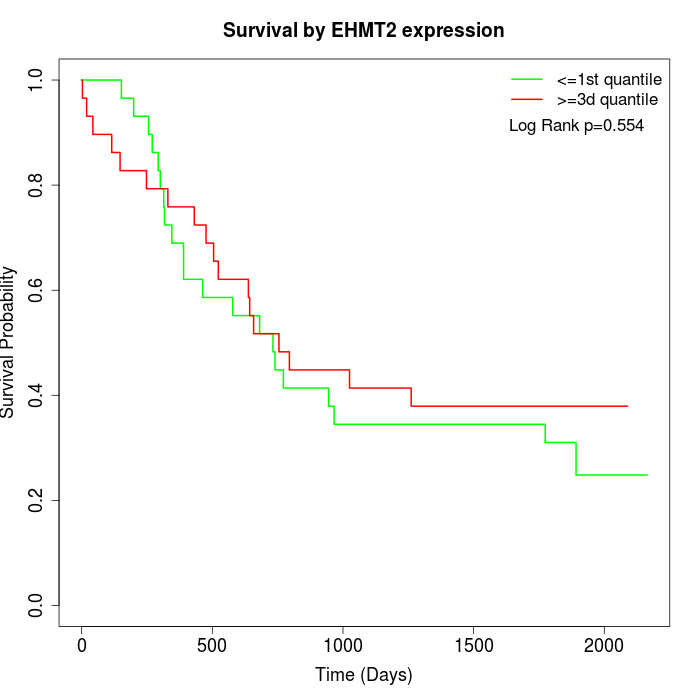
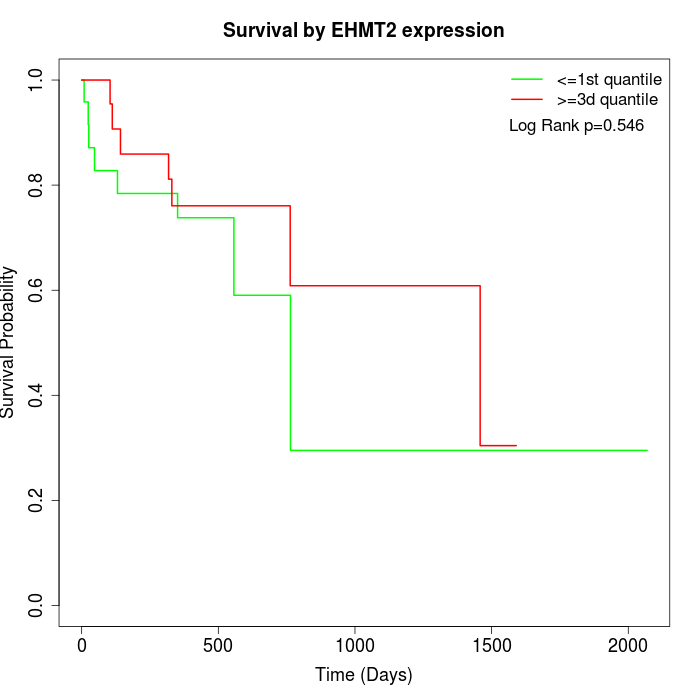
Survival by EHMT2 expression:
Note: Click image to view full size file.
Copy number change of EHMT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EHMT2 | 10919 | 5 | 1 | 24 | |
| GSE20123 | EHMT2 | 10919 | 5 | 1 | 24 | |
| GSE43470 | EHMT2 | 10919 | 5 | 1 | 37 | |
| GSE46452 | EHMT2 | 10919 | 1 | 10 | 48 | |
| GSE47630 | EHMT2 | 10919 | 7 | 4 | 29 | |
| GSE54993 | EHMT2 | 10919 | 2 | 1 | 67 | |
| GSE54994 | EHMT2 | 10919 | 11 | 4 | 38 | |
| GSE60625 | EHMT2 | 10919 | 0 | 1 | 10 | |
| GSE74703 | EHMT2 | 10919 | 5 | 0 | 31 | |
| GSE74704 | EHMT2 | 10919 | 2 | 0 | 18 | |
| TCGA | EHMT2 | 10919 | 16 | 16 | 64 |
Total number of gains: 59; Total number of losses: 39; Total Number of normals: 390.
Somatic mutations of EHMT2:
Generating mutation plots.
Highly correlated genes for EHMT2:
Showing top 20/756 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EHMT2 | CRTC2 | 0.8356 | 3 | 0 | 3 |
| EHMT2 | SPSB2 | 0.825065 | 3 | 0 | 3 |
| EHMT2 | ZNF641 | 0.795676 | 3 | 0 | 3 |
| EHMT2 | COMMD7 | 0.793849 | 3 | 0 | 3 |
| EHMT2 | C12orf73 | 0.793161 | 3 | 0 | 3 |
| EHMT2 | ZNF628 | 0.792106 | 3 | 0 | 3 |
| EHMT2 | WDR81 | 0.781476 | 3 | 0 | 3 |
| EHMT2 | ZNF324B | 0.775488 | 3 | 0 | 3 |
| EHMT2 | RRP36 | 0.775405 | 3 | 0 | 3 |
| EHMT2 | MLLT6 | 0.762201 | 3 | 0 | 3 |
| EHMT2 | ZNF680 | 0.759479 | 3 | 0 | 3 |
| EHMT2 | SSBP4 | 0.754539 | 4 | 0 | 3 |
| EHMT2 | MRPS9 | 0.753228 | 3 | 0 | 3 |
| EHMT2 | FCHO1 | 0.747325 | 4 | 0 | 4 |
| EHMT2 | GTPBP6 | 0.745646 | 3 | 0 | 3 |
| EHMT2 | SIX5 | 0.744665 | 3 | 0 | 3 |
| EHMT2 | PDCD2L | 0.739755 | 3 | 0 | 3 |
| EHMT2 | BATF3 | 0.739547 | 3 | 0 | 3 |
| EHMT2 | EFTUD2 | 0.738635 | 4 | 0 | 4 |
| EHMT2 | INPP5B | 0.73811 | 4 | 0 | 4 |
For details and further investigation, click here