| Full name: filamin B | Alias Symbol: TAP|TABP|ABP-278|FH1 | ||
| Type: protein-coding gene | Cytoband: 3p14.3 | ||
| Entrez ID: 2317 | HGNC ID: HGNC:3755 | Ensembl Gene: ENSG00000136068 | OMIM ID: 603381 |
| Drug and gene relationship at DGIdb | |||
FLNB involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04510 | Focal adhesion | |
| hsa05132 | Salmonella infection | |
| hsa05205 | Proteoglycans in cancer |
Expression of FLNB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FLNB | 2317 | 208614_s_at | 0.3615 | 0.5703 | |
| GSE20347 | FLNB | 2317 | 208614_s_at | 0.2080 | 0.3813 | |
| GSE23400 | FLNB | 2317 | 208614_s_at | 0.2348 | 0.0023 | |
| GSE26886 | FLNB | 2317 | 208614_s_at | 0.2142 | 0.3762 | |
| GSE29001 | FLNB | 2317 | 208614_s_at | 0.4074 | 0.0647 | |
| GSE38129 | FLNB | 2317 | 208614_s_at | 0.1978 | 0.2061 | |
| GSE45670 | FLNB | 2317 | 208614_s_at | 0.3993 | 0.0437 | |
| GSE53622 | FLNB | 2317 | 69147 | 0.1191 | 0.2194 | |
| GSE53624 | FLNB | 2317 | 69147 | 0.2038 | 0.0024 | |
| GSE63941 | FLNB | 2317 | 208614_s_at | -0.6856 | 0.2451 | |
| GSE77861 | FLNB | 2317 | 208614_s_at | 0.4637 | 0.0802 | |
| GSE97050 | FLNB | 2317 | A_23_P211878 | 0.1403 | 0.4801 | |
| SRP007169 | FLNB | 2317 | RNAseq | 0.5723 | 0.0949 | |
| SRP008496 | FLNB | 2317 | RNAseq | 0.6345 | 0.0102 | |
| SRP064894 | FLNB | 2317 | RNAseq | 0.4827 | 0.0103 | |
| SRP133303 | FLNB | 2317 | RNAseq | 0.3343 | 0.1598 | |
| SRP159526 | FLNB | 2317 | RNAseq | 0.1500 | 0.6321 | |
| SRP193095 | FLNB | 2317 | RNAseq | 0.3507 | 0.0973 | |
| SRP219564 | FLNB | 2317 | RNAseq | -0.3606 | 0.1808 | |
| TCGA | FLNB | 2317 | RNAseq | -0.0607 | 0.3470 |
Upregulated datasets: 0; Downregulated datasets: 0.
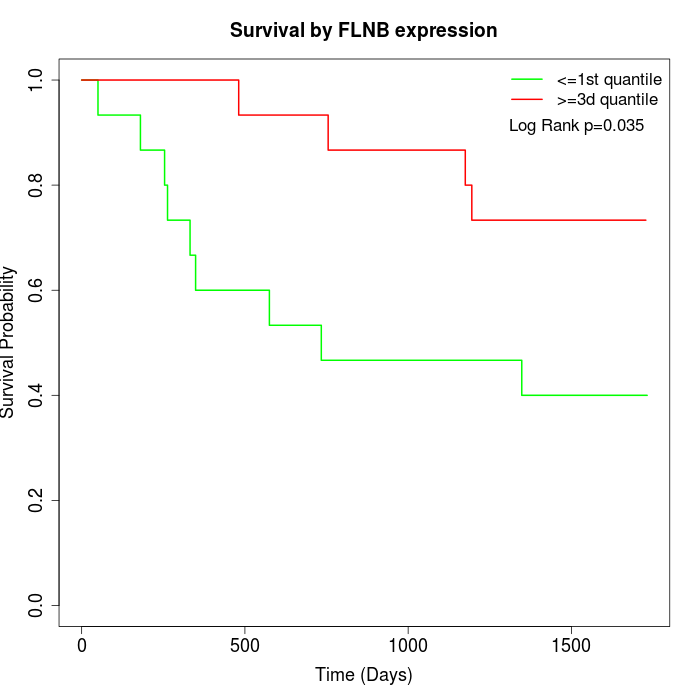
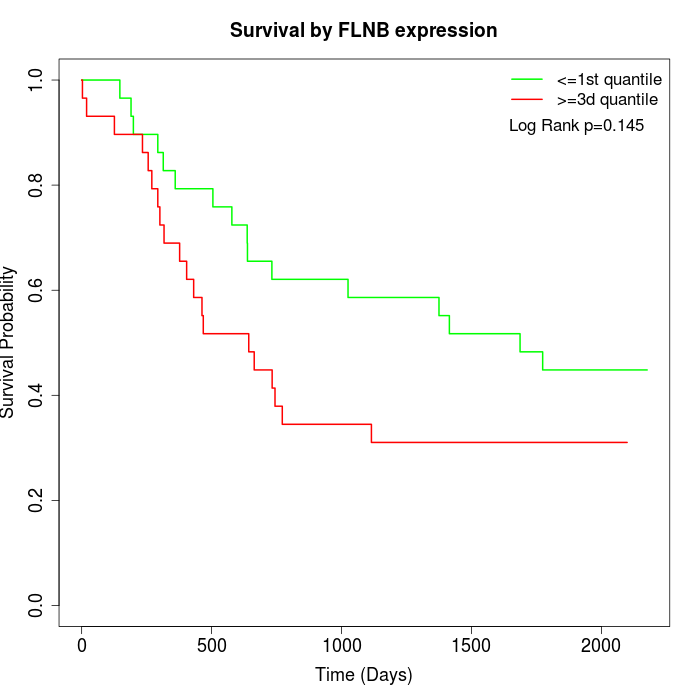
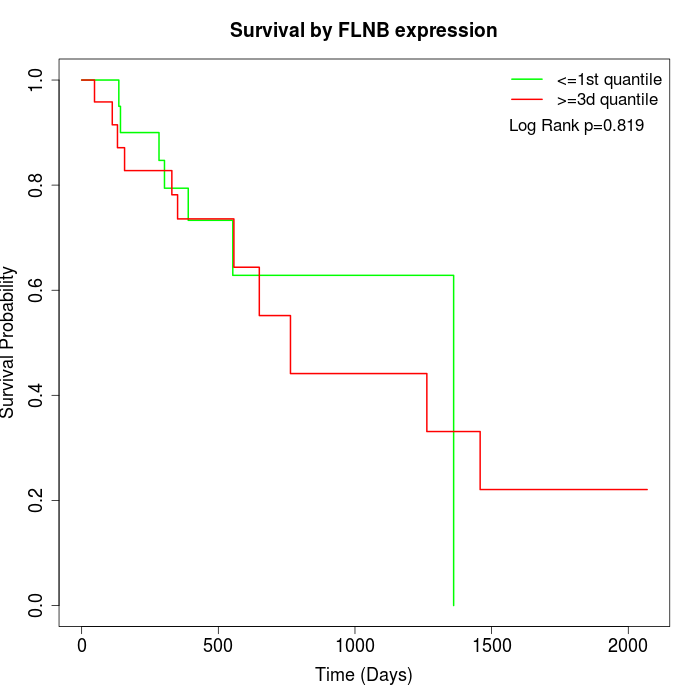
Survival by FLNB expression:
Note: Click image to view full size file.
Copy number change of FLNB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FLNB | 2317 | 0 | 17 | 13 | |
| GSE20123 | FLNB | 2317 | 0 | 18 | 12 | |
| GSE43470 | FLNB | 2317 | 0 | 17 | 26 | |
| GSE46452 | FLNB | 2317 | 2 | 17 | 40 | |
| GSE47630 | FLNB | 2317 | 1 | 24 | 15 | |
| GSE54993 | FLNB | 2317 | 6 | 2 | 62 | |
| GSE54994 | FLNB | 2317 | 0 | 32 | 21 | |
| GSE60625 | FLNB | 2317 | 5 | 0 | 6 | |
| GSE74703 | FLNB | 2317 | 0 | 14 | 22 | |
| GSE74704 | FLNB | 2317 | 0 | 11 | 9 | |
| TCGA | FLNB | 2317 | 0 | 79 | 17 |
Total number of gains: 14; Total number of losses: 231; Total Number of normals: 243.
Somatic mutations of FLNB:
Generating mutation plots.
Highly correlated genes for FLNB:
Showing top 20/671 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FLNB | GAA | 0.81244 | 3 | 0 | 3 |
| FLNB | SLC2A9 | 0.784389 | 3 | 0 | 3 |
| FLNB | USP40 | 0.770434 | 3 | 0 | 3 |
| FLNB | NOL9 | 0.765281 | 3 | 0 | 3 |
| FLNB | CERCAM | 0.754075 | 3 | 0 | 3 |
| FLNB | PLXND1 | 0.750788 | 3 | 0 | 3 |
| FLNB | NRM | 0.748476 | 3 | 0 | 3 |
| FLNB | SIVA1 | 0.741727 | 3 | 0 | 3 |
| FLNB | WDR54 | 0.738807 | 4 | 0 | 4 |
| FLNB | TMEM173 | 0.736881 | 3 | 0 | 3 |
| FLNB | TMEM17 | 0.729674 | 3 | 0 | 3 |
| FLNB | PLEKHB2 | 0.72648 | 3 | 0 | 3 |
| FLNB | NINJ1 | 0.724888 | 3 | 0 | 3 |
| FLNB | RBM14 | 0.723915 | 3 | 0 | 3 |
| FLNB | VPS39 | 0.723153 | 3 | 0 | 3 |
| FLNB | SH3RF3 | 0.722575 | 3 | 0 | 3 |
| FLNB | GSR | 0.72234 | 3 | 0 | 3 |
| FLNB | CCDC51 | 0.721586 | 3 | 0 | 3 |
| FLNB | SEL1L3 | 0.720547 | 4 | 0 | 4 |
| FLNB | SLC2A3 | 0.720269 | 3 | 0 | 3 |
For details and further investigation, click here