| Full name: RNA binding motif protein 14 | Alias Symbol: SIP|SYTIP1|COAA|DKFZp779J0927 | ||
| Type: protein-coding gene | Cytoband: 11q13.2 | ||
| Entrez ID: 10432 | HGNC ID: HGNC:14219 | Ensembl Gene: ENSG00000239306 | OMIM ID: 612409 |
| Drug and gene relationship at DGIdb | |||
Expression of RBM14:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RBM14 | 10432 | 204178_s_at | 0.6581 | 0.1286 | |
| GSE20347 | RBM14 | 10432 | 204178_s_at | 0.4698 | 0.0000 | |
| GSE23400 | RBM14 | 10432 | 204178_s_at | 0.3409 | 0.0000 | |
| GSE26886 | RBM14 | 10432 | 204178_s_at | -0.3073 | 0.2190 | |
| GSE29001 | RBM14 | 10432 | 204178_s_at | 0.3936 | 0.2441 | |
| GSE38129 | RBM14 | 10432 | 204178_s_at | 0.4801 | 0.0000 | |
| GSE45670 | RBM14 | 10432 | 204178_s_at | 0.6054 | 0.0003 | |
| GSE53622 | RBM14 | 10432 | 31416 | 0.4038 | 0.0000 | |
| GSE53624 | RBM14 | 10432 | 31416 | 0.3755 | 0.0000 | |
| GSE63941 | RBM14 | 10432 | 204178_s_at | 0.7320 | 0.0294 | |
| GSE77861 | RBM14 | 10432 | 204178_s_at | 0.4158 | 0.0991 | |
| GSE97050 | RBM14 | 10432 | A_23_P150255 | 0.2602 | 0.2199 | |
| SRP007169 | RBM14 | 10432 | RNAseq | 0.3250 | 0.3369 | |
| SRP008496 | RBM14 | 10432 | RNAseq | 0.3282 | 0.1756 | |
| SRP064894 | RBM14 | 10432 | RNAseq | 0.3030 | 0.0850 | |
| SRP133303 | RBM14 | 10432 | RNAseq | 0.2795 | 0.0509 | |
| SRP159526 | RBM14 | 10432 | RNAseq | 0.4193 | 0.1305 | |
| SRP193095 | RBM14 | 10432 | RNAseq | 0.3237 | 0.0069 | |
| SRP219564 | RBM14 | 10432 | RNAseq | 0.2475 | 0.2060 | |
| TCGA | RBM14 | 10432 | RNAseq | 0.0769 | 0.0912 |
Upregulated datasets: 0; Downregulated datasets: 0.
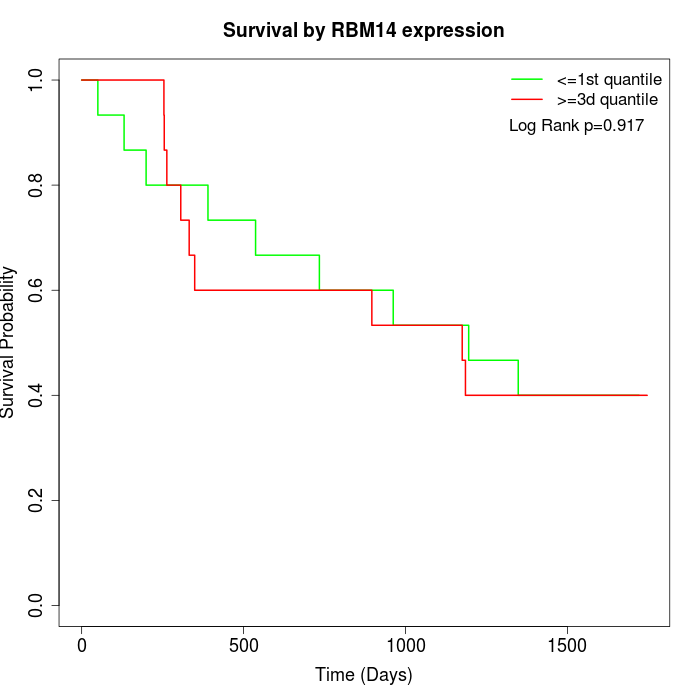
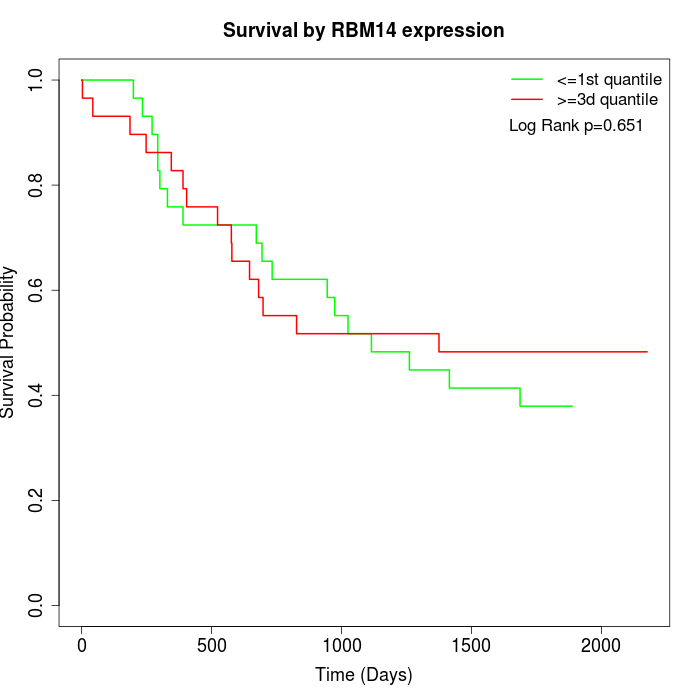
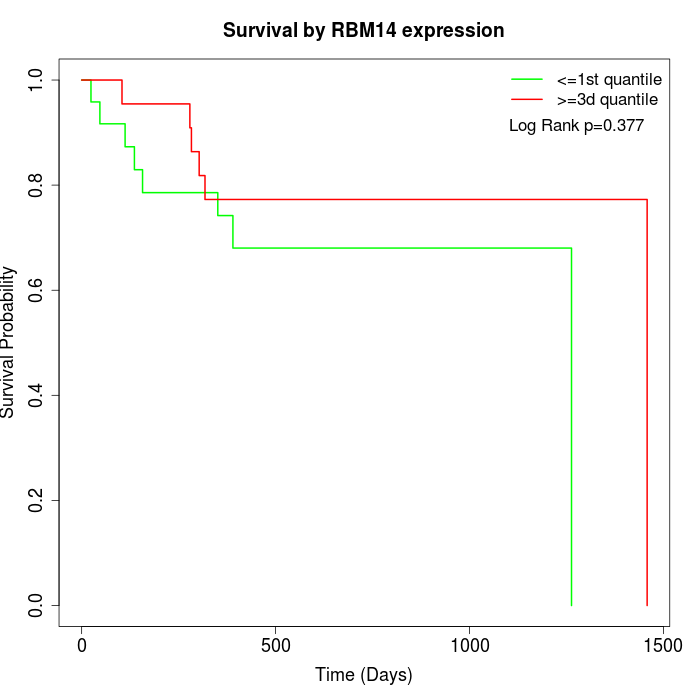
Survival by RBM14 expression:
Note: Click image to view full size file.
Copy number change of RBM14:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RBM14 | 10432 | 9 | 4 | 17 | |
| GSE20123 | RBM14 | 10432 | 9 | 4 | 17 | |
| GSE43470 | RBM14 | 10432 | 6 | 1 | 36 | |
| GSE46452 | RBM14 | 10432 | 12 | 3 | 44 | |
| GSE47630 | RBM14 | 10432 | 9 | 4 | 27 | |
| GSE54993 | RBM14 | 10432 | 3 | 0 | 67 | |
| GSE54994 | RBM14 | 10432 | 10 | 4 | 39 | |
| GSE60625 | RBM14 | 10432 | 0 | 3 | 8 | |
| GSE74703 | RBM14 | 10432 | 4 | 0 | 32 | |
| GSE74704 | RBM14 | 10432 | 7 | 2 | 11 | |
| TCGA | RBM14 | 10432 | 27 | 6 | 63 |
Total number of gains: 96; Total number of losses: 31; Total Number of normals: 361.
Somatic mutations of RBM14:
Generating mutation plots.
Highly correlated genes for RBM14:
Showing top 20/1189 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RBM14 | OAF | 0.81314 | 3 | 0 | 3 |
| RBM14 | ABCD4 | 0.76593 | 3 | 0 | 3 |
| RBM14 | POLDIP3 | 0.760186 | 3 | 0 | 3 |
| RBM14 | SLC39A3 | 0.755938 | 3 | 0 | 3 |
| RBM14 | DEDD | 0.725072 | 4 | 0 | 4 |
| RBM14 | FUCA2 | 0.724051 | 3 | 0 | 3 |
| RBM14 | FLNB | 0.723915 | 3 | 0 | 3 |
| RBM14 | PKN1 | 0.706102 | 4 | 0 | 3 |
| RBM14 | CD99L2 | 0.704893 | 3 | 0 | 3 |
| RBM14 | TCF7L1 | 0.704359 | 3 | 0 | 3 |
| RBM14 | BEST1 | 0.699823 | 3 | 0 | 3 |
| RBM14 | LSM4 | 0.69339 | 9 | 0 | 9 |
| RBM14 | DRAP1 | 0.692971 | 3 | 0 | 3 |
| RBM14 | ZC3HC1 | 0.691332 | 5 | 0 | 4 |
| RBM14 | SUPT16H | 0.691285 | 8 | 0 | 8 |
| RBM14 | E2F6 | 0.689779 | 8 | 0 | 8 |
| RBM14 | ELP2 | 0.688273 | 3 | 0 | 3 |
| RBM14 | APLNR | 0.684584 | 4 | 0 | 4 |
| RBM14 | DOCK6 | 0.684164 | 4 | 0 | 4 |
| RBM14 | ANKRD54 | 0.679681 | 5 | 0 | 5 |
For details and further investigation, click here