| Full name: plexin D1 | Alias Symbol: KIAA0620 | ||
| Type: protein-coding gene | Cytoband: 3q22.1 | ||
| Entrez ID: 23129 | HGNC ID: HGNC:9107 | Ensembl Gene: ENSG00000004399 | OMIM ID: 604282 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PLXND1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PLXND1 | 23129 | 38671_at | 0.7569 | 0.1173 | |
| GSE20347 | PLXND1 | 23129 | 38671_at | 0.7060 | 0.0000 | |
| GSE23400 | PLXND1 | 23129 | 38671_at | 0.3917 | 0.0000 | |
| GSE26886 | PLXND1 | 23129 | 38671_at | 0.7533 | 0.0003 | |
| GSE29001 | PLXND1 | 23129 | 38671_at | 1.0503 | 0.0025 | |
| GSE38129 | PLXND1 | 23129 | 38671_at | 0.3283 | 0.1132 | |
| GSE45670 | PLXND1 | 23129 | 38671_at | -0.0205 | 0.9468 | |
| GSE53622 | PLXND1 | 23129 | 164140 | 0.3022 | 0.0004 | |
| GSE53624 | PLXND1 | 23129 | 164140 | 0.6930 | 0.0000 | |
| GSE63941 | PLXND1 | 23129 | 38671_at | -1.7326 | 0.0039 | |
| GSE77861 | PLXND1 | 23129 | 38671_at | 0.6536 | 0.0038 | |
| GSE97050 | PLXND1 | 23129 | A_24_P376391 | 0.4852 | 0.1729 | |
| SRP007169 | PLXND1 | 23129 | RNAseq | 3.0290 | 0.0000 | |
| SRP008496 | PLXND1 | 23129 | RNAseq | 2.8140 | 0.0000 | |
| SRP064894 | PLXND1 | 23129 | RNAseq | 1.3785 | 0.0000 | |
| SRP133303 | PLXND1 | 23129 | RNAseq | 0.3478 | 0.1533 | |
| SRP159526 | PLXND1 | 23129 | RNAseq | 0.6535 | 0.0044 | |
| SRP193095 | PLXND1 | 23129 | RNAseq | 1.5798 | 0.0000 | |
| SRP219564 | PLXND1 | 23129 | RNAseq | 1.0444 | 0.0053 | |
| TCGA | PLXND1 | 23129 | RNAseq | 0.0760 | 0.3498 |
Upregulated datasets: 6; Downregulated datasets: 1.
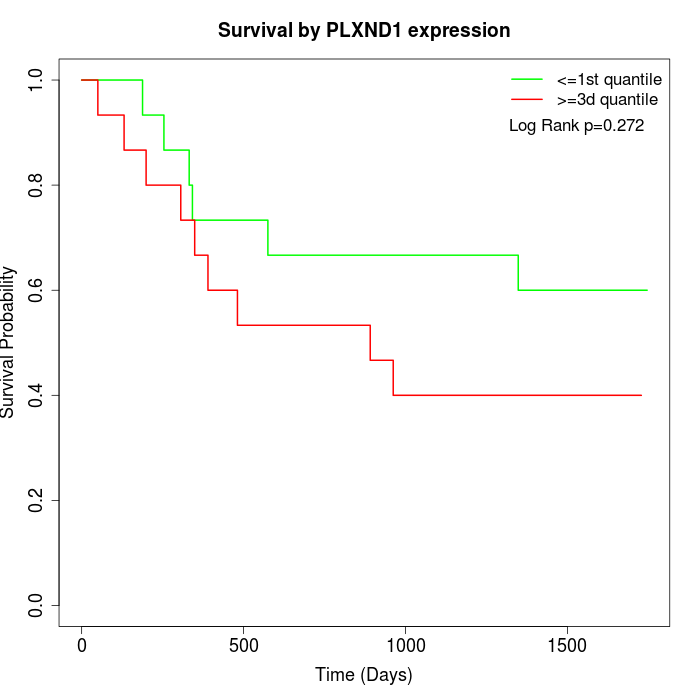
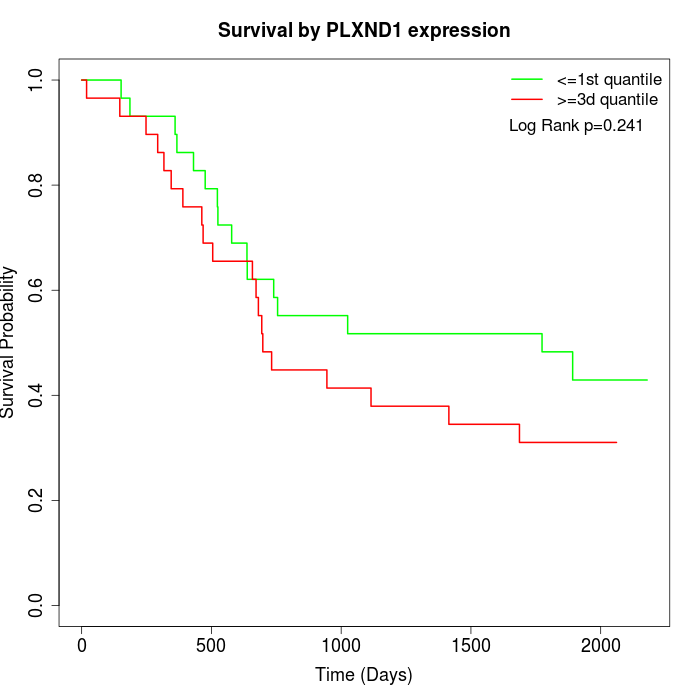
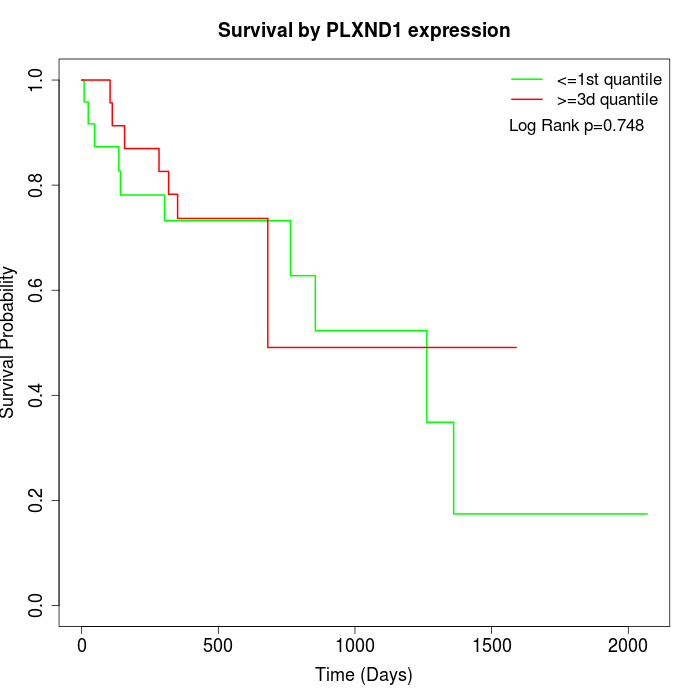
Survival by PLXND1 expression:
Note: Click image to view full size file.
Copy number change of PLXND1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PLXND1 | 23129 | 17 | 0 | 13 | |
| GSE20123 | PLXND1 | 23129 | 17 | 0 | 13 | |
| GSE43470 | PLXND1 | 23129 | 19 | 0 | 24 | |
| GSE46452 | PLXND1 | 23129 | 14 | 5 | 40 | |
| GSE47630 | PLXND1 | 23129 | 18 | 3 | 19 | |
| GSE54993 | PLXND1 | 23129 | 2 | 8 | 60 | |
| GSE54994 | PLXND1 | 23129 | 32 | 3 | 18 | |
| GSE60625 | PLXND1 | 23129 | 0 | 6 | 5 | |
| GSE74703 | PLXND1 | 23129 | 17 | 0 | 19 | |
| GSE74704 | PLXND1 | 23129 | 13 | 0 | 7 | |
| TCGA | PLXND1 | 23129 | 59 | 5 | 32 |
Total number of gains: 208; Total number of losses: 30; Total Number of normals: 250.
Somatic mutations of PLXND1:
Generating mutation plots.
Highly correlated genes for PLXND1:
Showing top 20/1399 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PLXND1 | TMC8 | 0.859573 | 3 | 0 | 3 |
| PLXND1 | MMP2 | 0.774152 | 10 | 0 | 10 |
| PLXND1 | PCOLCE | 0.765992 | 12 | 0 | 12 |
| PLXND1 | SLC39A3 | 0.756196 | 3 | 0 | 3 |
| PLXND1 | COL6A3 | 0.751876 | 12 | 0 | 12 |
| PLXND1 | FLNB | 0.750788 | 3 | 0 | 3 |
| PLXND1 | PDGFRB | 0.748464 | 11 | 0 | 10 |
| PLXND1 | GAS2L1 | 0.746689 | 3 | 0 | 3 |
| PLXND1 | LRRC8C | 0.74584 | 3 | 0 | 3 |
| PLXND1 | VCAN | 0.74151 | 12 | 0 | 12 |
| PLXND1 | SPIRE1 | 0.740708 | 3 | 0 | 3 |
| PLXND1 | PEAR1 | 0.739253 | 4 | 0 | 4 |
| PLXND1 | NACC1 | 0.738638 | 3 | 0 | 3 |
| PLXND1 | SPARC | 0.734799 | 12 | 0 | 11 |
| PLXND1 | CTSK | 0.73307 | 12 | 0 | 12 |
| PLXND1 | OLFML2B | 0.732861 | 12 | 0 | 12 |
| PLXND1 | GDF11 | 0.732 | 3 | 0 | 3 |
| PLXND1 | RNF217 | 0.730035 | 3 | 0 | 3 |
| PLXND1 | AEBP1 | 0.724594 | 12 | 0 | 12 |
| PLXND1 | TRIM47 | 0.72444 | 4 | 0 | 4 |
For details and further investigation, click here