| Full name: polypeptide N-acetylgalactosaminyltransferase 9 | Alias Symbol: GALNAC-T9 | ||
| Type: protein-coding gene | Cytoband: 12q24.33 | ||
| Entrez ID: 50614 | HGNC ID: HGNC:4131 | Ensembl Gene: ENSG00000182870 | OMIM ID: 606251 |
| Drug and gene relationship at DGIdb | |||
Expression of GALNT9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GALNT9 | 50614 | 229451_at | -0.0861 | 0.6890 | |
| GSE26886 | GALNT9 | 50614 | 229451_at | -0.0923 | 0.5239 | |
| GSE45670 | GALNT9 | 50614 | 229451_at | -0.0032 | 0.9793 | |
| GSE53622 | GALNT9 | 50614 | 16890 | -1.6432 | 0.0000 | |
| GSE53624 | GALNT9 | 50614 | 16890 | -1.6828 | 0.0000 | |
| GSE63941 | GALNT9 | 50614 | 229451_at | 0.0007 | 0.9975 | |
| GSE77861 | GALNT9 | 50614 | 229451_at | -0.0930 | 0.3577 | |
| GSE97050 | GALNT9 | A_19_P00322242 | -0.2740 | 0.5092 | ||
| TCGA | GALNT9 | 50614 | RNAseq | -1.4265 | 0.0914 |
Upregulated datasets: 0; Downregulated datasets: 2.
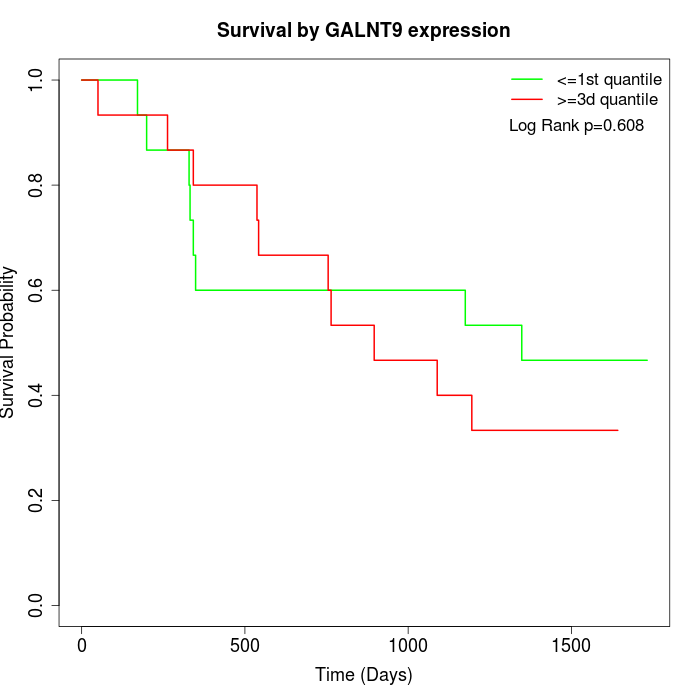
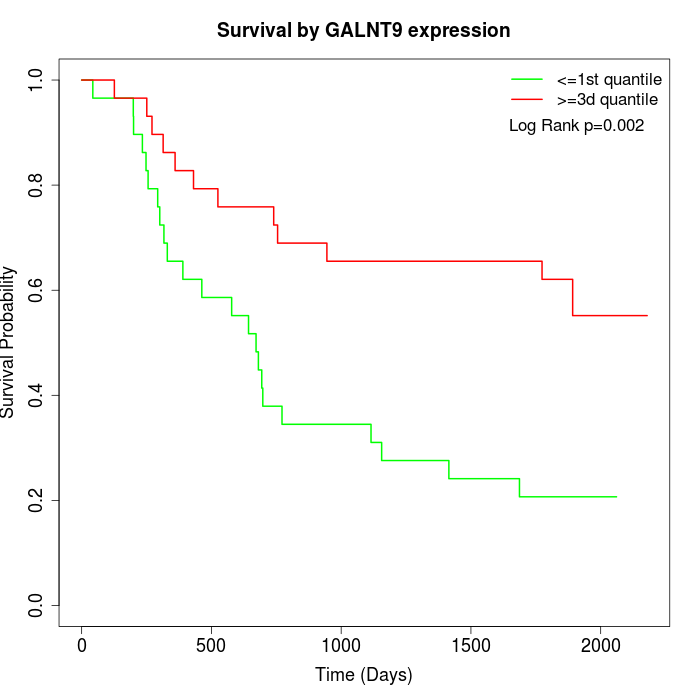
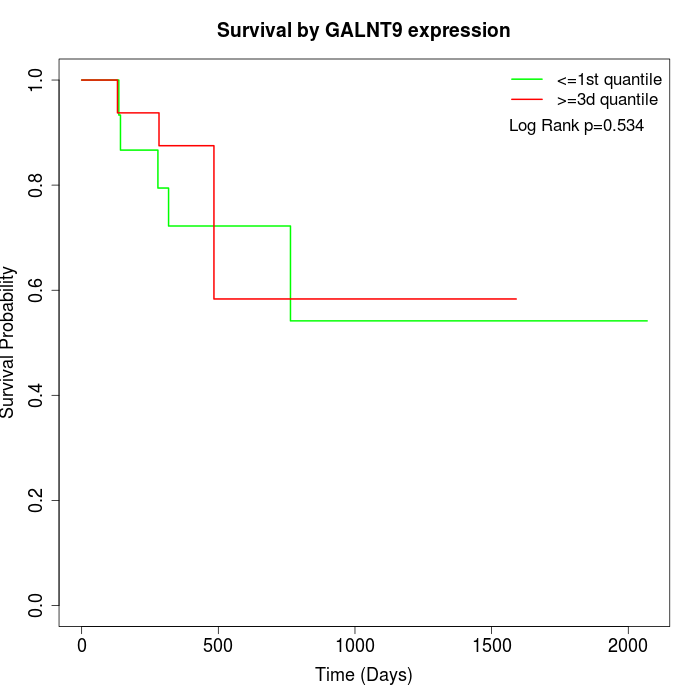
Survival by GALNT9 expression:
Note: Click image to view full size file.
Copy number change of GALNT9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GALNT9 | 50614 | 4 | 5 | 21 | |
| GSE20123 | GALNT9 | 50614 | 4 | 5 | 21 | |
| GSE43470 | GALNT9 | 50614 | 3 | 3 | 37 | |
| GSE46452 | GALNT9 | 50614 | 8 | 3 | 48 | |
| GSE47630 | GALNT9 | 50614 | 9 | 3 | 28 | |
| GSE54993 | GALNT9 | 50614 | 0 | 6 | 64 | |
| GSE54994 | GALNT9 | 50614 | 4 | 8 | 41 | |
| GSE60625 | GALNT9 | 50614 | 0 | 0 | 11 | |
| GSE74703 | GALNT9 | 50614 | 2 | 3 | 31 | |
| GSE74704 | GALNT9 | 50614 | 2 | 3 | 15 | |
| TCGA | GALNT9 | 50614 | 22 | 15 | 59 |
Total number of gains: 58; Total number of losses: 54; Total Number of normals: 376.
Somatic mutations of GALNT9:
Generating mutation plots.
Highly correlated genes for GALNT9:
Showing top 20/582 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GALNT9 | KRTAP20-1 | 0.897439 | 3 | 0 | 3 |
| GALNT9 | OR2Z1 | 0.885894 | 3 | 0 | 3 |
| GALNT9 | MFSD2B | 0.869514 | 3 | 0 | 3 |
| GALNT9 | OR10G8 | 0.861621 | 3 | 0 | 3 |
| GALNT9 | FAM163B | 0.830599 | 3 | 0 | 3 |
| GALNT9 | CLDN24 | 0.825594 | 3 | 0 | 3 |
| GALNT9 | LINC01141 | 0.805429 | 3 | 0 | 3 |
| GALNT9 | SLC25A47 | 0.804945 | 3 | 0 | 3 |
| GALNT9 | BTBD9 | 0.783287 | 3 | 0 | 3 |
| GALNT9 | ARID5A | 0.779864 | 3 | 0 | 3 |
| GALNT9 | UPB1 | 0.779585 | 4 | 0 | 4 |
| GALNT9 | SLC25A48 | 0.778674 | 3 | 0 | 3 |
| GALNT9 | TEN1 | 0.774966 | 3 | 0 | 3 |
| GALNT9 | PKD2L1 | 0.774665 | 5 | 0 | 5 |
| GALNT9 | HES5 | 0.774226 | 3 | 0 | 3 |
| GALNT9 | C16orf89 | 0.762808 | 3 | 0 | 3 |
| GALNT9 | BARHL2 | 0.759015 | 3 | 0 | 3 |
| GALNT9 | CD164L2 | 0.749739 | 3 | 0 | 3 |
| GALNT9 | DEF6 | 0.747222 | 3 | 0 | 3 |
| GALNT9 | ZMYND10 | 0.745362 | 3 | 0 | 3 |
For details and further investigation, click here