| Full name: hes family bHLH transcription factor 5 | Alias Symbol: bHLHb38 | ||
| Type: protein-coding gene | Cytoband: 1p36.32 | ||
| Entrez ID: 388585 | HGNC ID: HGNC:19764 | Ensembl Gene: ENSG00000197921 | OMIM ID: 607348 |
| Drug and gene relationship at DGIdb | |||
HES5 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04330 | Notch signaling pathway |
Expression of HES5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HES5 | 388585 | 239230_at | 0.3557 | 0.4781 | |
| GSE26886 | HES5 | 388585 | 239230_at | -0.5739 | 0.0325 | |
| GSE45670 | HES5 | 388585 | 239230_at | 0.4659 | 0.0375 | |
| GSE53622 | HES5 | 388585 | 54528 | -1.8725 | 0.0000 | |
| GSE53624 | HES5 | 388585 | 54528 | -2.2238 | 0.0000 | |
| GSE63941 | HES5 | 388585 | 239230_at | 0.4751 | 0.1050 | |
| GSE77861 | HES5 | 388585 | 239230_at | 0.0682 | 0.8209 | |
| GSE97050 | HES5 | 388585 | A_32_P234184 | -1.4465 | 0.1270 | |
| SRP133303 | HES5 | 388585 | RNAseq | 0.1250 | 0.7115 | |
| TCGA | HES5 | 388585 | RNAseq | 0.5551 | 0.2270 |
Upregulated datasets: 0; Downregulated datasets: 2.
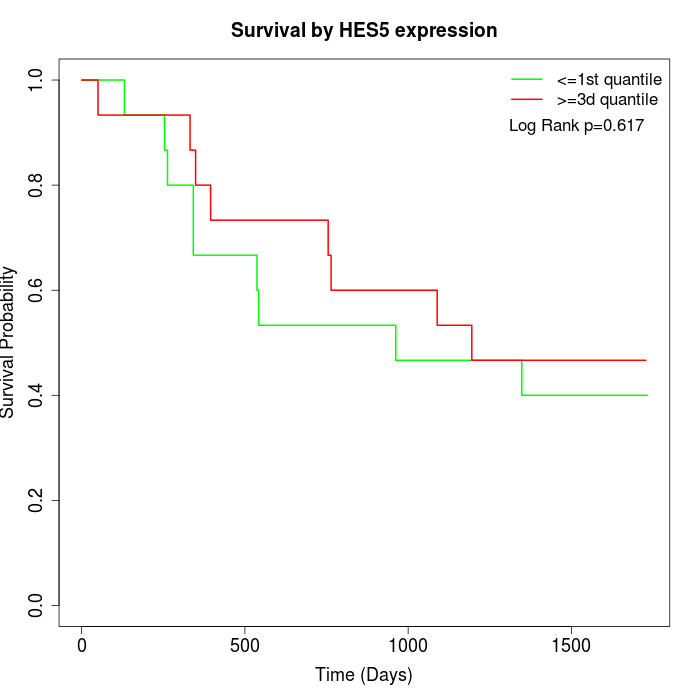
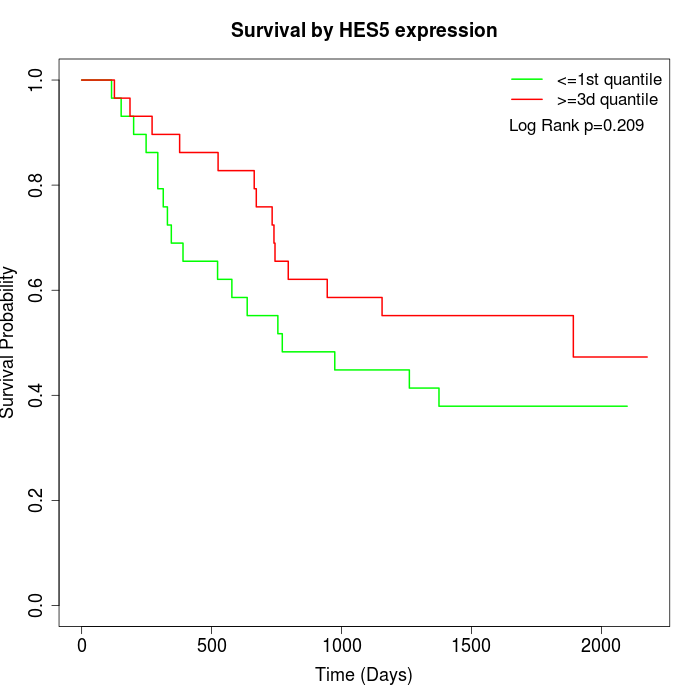
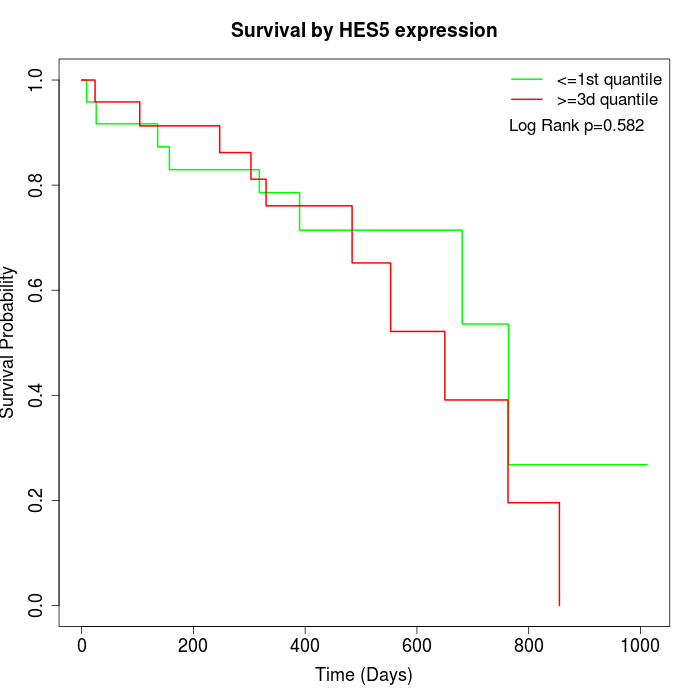
Survival by HES5 expression:
Note: Click image to view full size file.
Copy number change of HES5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HES5 | 388585 | 4 | 6 | 20 | |
| GSE20123 | HES5 | 388585 | 4 | 5 | 21 | |
| GSE43470 | HES5 | 388585 | 2 | 5 | 36 | |
| GSE46452 | HES5 | 388585 | 7 | 1 | 51 | |
| GSE47630 | HES5 | 388585 | 8 | 4 | 28 | |
| GSE54993 | HES5 | 388585 | 3 | 2 | 65 | |
| GSE54994 | HES5 | 388585 | 15 | 2 | 36 | |
| GSE60625 | HES5 | 388585 | 0 | 0 | 11 | |
| GSE74703 | HES5 | 388585 | 2 | 3 | 31 | |
| GSE74704 | HES5 | 388585 | 4 | 0 | 16 | |
| TCGA | HES5 | 388585 | 14 | 20 | 62 |
Total number of gains: 63; Total number of losses: 48; Total Number of normals: 377.
Somatic mutations of HES5:
Generating mutation plots.
Highly correlated genes for HES5:
Showing top 20/518 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HES5 | FGF5 | 0.906329 | 3 | 0 | 3 |
| HES5 | FAM163B | 0.881826 | 3 | 0 | 3 |
| HES5 | NCCRP1 | 0.860246 | 3 | 0 | 3 |
| HES5 | MUC22 | 0.829179 | 3 | 0 | 3 |
| HES5 | KRTAP20-1 | 0.823629 | 3 | 0 | 3 |
| HES5 | ETHE1 | 0.822218 | 3 | 0 | 3 |
| HES5 | CYSRT1 | 0.818572 | 3 | 0 | 3 |
| HES5 | MFSD2B | 0.816192 | 3 | 0 | 3 |
| HES5 | OTOP3 | 0.815192 | 3 | 0 | 3 |
| HES5 | OR2Z1 | 0.814928 | 3 | 0 | 3 |
| HES5 | SPINK7 | 0.814253 | 4 | 0 | 4 |
| HES5 | HMGB4 | 0.814056 | 3 | 0 | 3 |
| HES5 | KPRP | 0.811459 | 3 | 0 | 3 |
| HES5 | CRTAC1 | 0.805904 | 3 | 0 | 3 |
| HES5 | RNF222 | 0.803598 | 3 | 0 | 3 |
| HES5 | EPS8L2 | 0.801103 | 4 | 0 | 4 |
| HES5 | TMPRSS11A | 0.789955 | 3 | 0 | 3 |
| HES5 | PRSS27 | 0.782142 | 4 | 0 | 3 |
| HES5 | CRCT1 | 0.781513 | 4 | 0 | 4 |
| HES5 | KCNK7 | 0.779638 | 4 | 0 | 4 |
For details and further investigation, click here