| Full name: polypeptide N-acetylgalactosaminyltransferase like 5 | Alias Symbol: GalNAc-T5L | ||
| Type: protein-coding gene | Cytoband: 7q36.1 | ||
| Entrez ID: 168391 | HGNC ID: HGNC:21725 | Ensembl Gene: ENSG00000106648 | OMIM ID: 615133 |
| Drug and gene relationship at DGIdb | |||
Expression of GALNTL5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GALNTL5 | 168391 | 233150_at | -0.0626 | 0.7858 | |
| GSE26886 | GALNTL5 | 168391 | 233150_at | 0.0591 | 0.6007 | |
| GSE45670 | GALNTL5 | 168391 | 233150_at | 0.0064 | 0.9408 | |
| GSE53622 | GALNTL5 | 168391 | 7947 | 0.2470 | 0.2345 | |
| GSE53624 | GALNTL5 | 168391 | 61730 | 0.0863 | 0.3663 | |
| GSE63941 | GALNTL5 | 168391 | 233150_at | 0.1387 | 0.5994 | |
| GSE77861 | GALNTL5 | 168391 | 233150_at | -0.1332 | 0.1972 |
Upregulated datasets: 0; Downregulated datasets: 0.
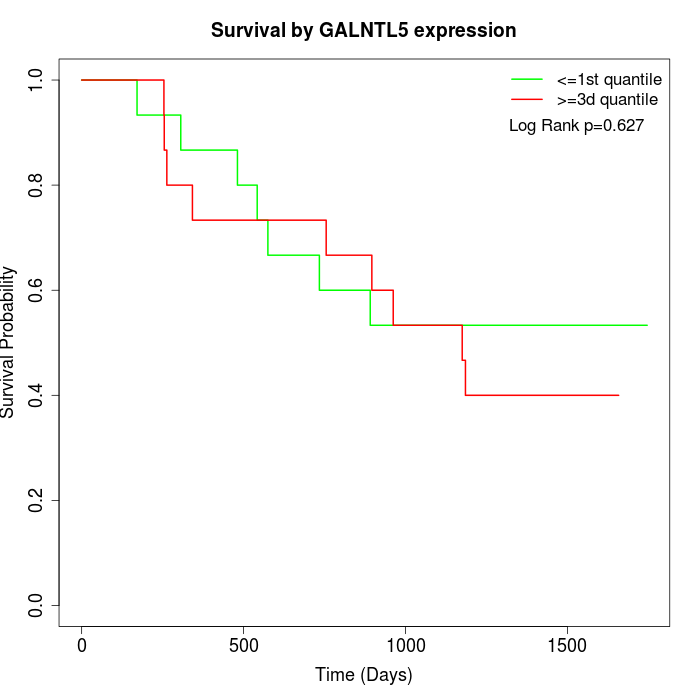
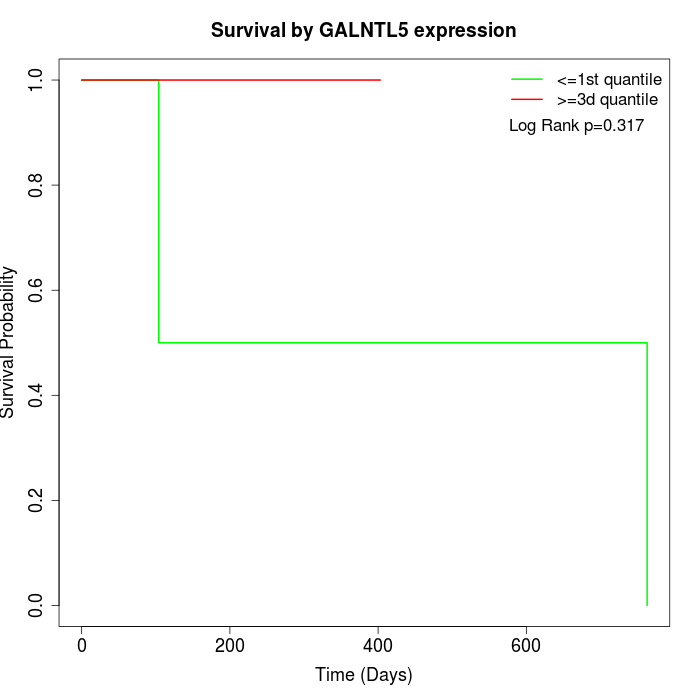
Survival by GALNTL5 expression:
Note: Click image to view full size file.
Copy number change of GALNTL5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GALNTL5 | 168391 | 2 | 4 | 24 | |
| GSE20123 | GALNTL5 | 168391 | 2 | 4 | 24 | |
| GSE43470 | GALNTL5 | 168391 | 2 | 5 | 36 | |
| GSE46452 | GALNTL5 | 168391 | 7 | 2 | 50 | |
| GSE47630 | GALNTL5 | 168391 | 6 | 9 | 25 | |
| GSE54993 | GALNTL5 | 168391 | 3 | 3 | 64 | |
| GSE54994 | GALNTL5 | 168391 | 6 | 11 | 36 | |
| GSE60625 | GALNTL5 | 168391 | 0 | 0 | 11 | |
| GSE74703 | GALNTL5 | 168391 | 2 | 4 | 30 | |
| GSE74704 | GALNTL5 | 168391 | 1 | 4 | 15 | |
| TCGA | GALNTL5 | 168391 | 26 | 27 | 43 |
Total number of gains: 57; Total number of losses: 73; Total Number of normals: 358.
Somatic mutations of GALNTL5:
Generating mutation plots.
Highly correlated genes for GALNTL5:
Showing top 20/69 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GALNTL5 | BFSP2-AS1 | 0.741511 | 3 | 0 | 3 |
| GALNTL5 | LINC00410 | 0.701183 | 3 | 0 | 3 |
| GALNTL5 | PRSS50 | 0.687822 | 3 | 0 | 3 |
| GALNTL5 | RTP3 | 0.679872 | 3 | 0 | 3 |
| GALNTL5 | PURB | 0.679409 | 3 | 0 | 3 |
| GALNTL5 | APOL5 | 0.672255 | 3 | 0 | 3 |
| GALNTL5 | ABCB5 | 0.665866 | 3 | 0 | 3 |
| GALNTL5 | KRTAP11-1 | 0.664959 | 3 | 0 | 3 |
| GALNTL5 | LINC01281 | 0.651225 | 3 | 0 | 3 |
| GALNTL5 | FGFR4 | 0.647405 | 3 | 0 | 3 |
| GALNTL5 | TEKT1 | 0.644313 | 3 | 0 | 3 |
| GALNTL5 | DLGAP2-AS1 | 0.642049 | 4 | 0 | 4 |
| GALNTL5 | DEFA4 | 0.636517 | 5 | 0 | 4 |
| GALNTL5 | MC3R | 0.636459 | 3 | 0 | 3 |
| GALNTL5 | SLC25A18 | 0.625901 | 3 | 0 | 3 |
| GALNTL5 | CRH | 0.607033 | 3 | 0 | 3 |
| GALNTL5 | PDYN | 0.604569 | 3 | 0 | 3 |
| GALNTL5 | KIRREL3-AS3 | 0.604343 | 3 | 0 | 3 |
| GALNTL5 | ZNF582-AS1 | 0.60158 | 4 | 0 | 3 |
| GALNTL5 | TAS2R16 | 0.59881 | 3 | 0 | 3 |
For details and further investigation, click here