| Full name: ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 | Alias Symbol: KIAA0041|CNT-B2 | ||
| Type: protein-coding gene | Cytoband: 3q29 | ||
| Entrez ID: 23527 | HGNC ID: HGNC:16469 | Ensembl Gene: ENSG00000114331 | OMIM ID: 607766 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of ACAP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ACAP2 | 23527 | 212476_at | -0.0729 | 0.9177 | |
| GSE20347 | ACAP2 | 23527 | 212476_at | -0.1388 | 0.4210 | |
| GSE23400 | ACAP2 | 23527 | 212476_at | -0.0403 | 0.7533 | |
| GSE26886 | ACAP2 | 23527 | 212476_at | 0.1926 | 0.4209 | |
| GSE29001 | ACAP2 | 23527 | 212476_at | -0.3076 | 0.5267 | |
| GSE38129 | ACAP2 | 23527 | 212476_at | 0.0095 | 0.9657 | |
| GSE45670 | ACAP2 | 23527 | 212476_at | -0.0557 | 0.8450 | |
| GSE53622 | ACAP2 | 23527 | 18615 | -0.0307 | 0.7565 | |
| GSE53624 | ACAP2 | 23527 | 18615 | -0.4719 | 0.0000 | |
| GSE63941 | ACAP2 | 23527 | 212476_at | -0.7098 | 0.2315 | |
| GSE77861 | ACAP2 | 23527 | 212476_at | -0.2337 | 0.6166 | |
| GSE97050 | ACAP2 | 23527 | A_24_P942002 | 0.1646 | 0.5554 | |
| SRP007169 | ACAP2 | 23527 | RNAseq | -0.2326 | 0.4574 | |
| SRP008496 | ACAP2 | 23527 | RNAseq | 0.0019 | 0.9918 | |
| SRP064894 | ACAP2 | 23527 | RNAseq | -0.5175 | 0.0731 | |
| SRP133303 | ACAP2 | 23527 | RNAseq | 0.1861 | 0.3504 | |
| SRP159526 | ACAP2 | 23527 | RNAseq | 0.3144 | 0.5096 | |
| SRP193095 | ACAP2 | 23527 | RNAseq | -0.2313 | 0.1494 | |
| SRP219564 | ACAP2 | 23527 | RNAseq | -0.5377 | 0.2623 | |
| TCGA | ACAP2 | 23527 | RNAseq | 0.1938 | 0.0017 |
Upregulated datasets: 0; Downregulated datasets: 0.
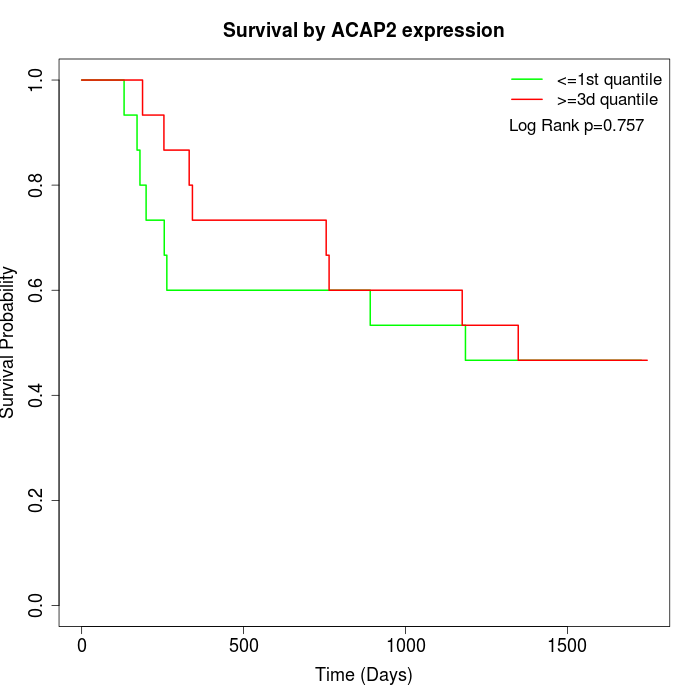
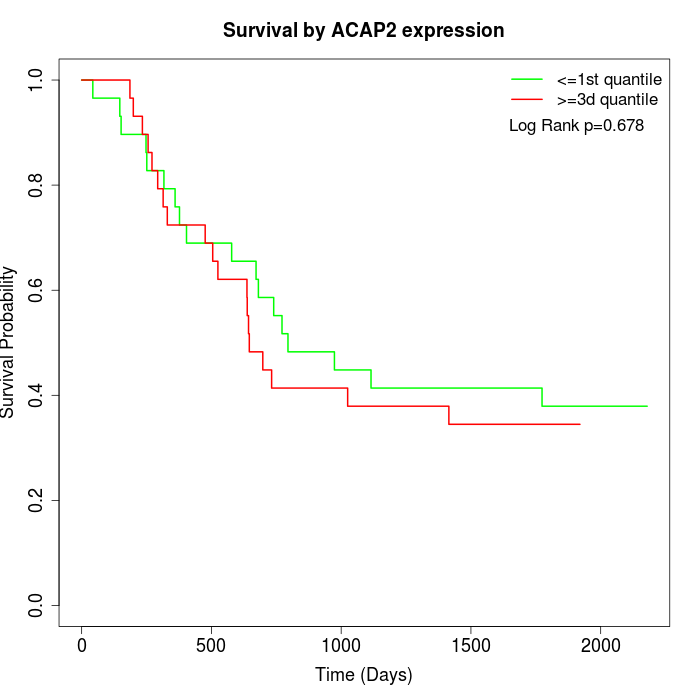
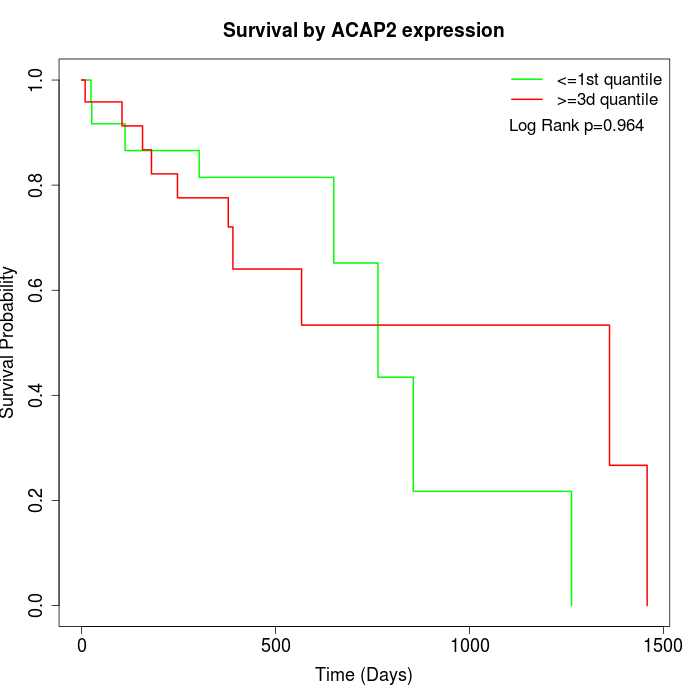
Survival by ACAP2 expression:
Note: Click image to view full size file.
Copy number change of ACAP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ACAP2 | 23527 | 22 | 0 | 8 | |
| GSE20123 | ACAP2 | 23527 | 22 | 0 | 8 | |
| GSE43470 | ACAP2 | 23527 | 23 | 0 | 20 | |
| GSE46452 | ACAP2 | 23527 | 19 | 1 | 39 | |
| GSE47630 | ACAP2 | 23527 | 27 | 2 | 11 | |
| GSE54993 | ACAP2 | 23527 | 1 | 22 | 47 | |
| GSE54994 | ACAP2 | 23527 | 41 | 0 | 12 | |
| GSE60625 | ACAP2 | 23527 | 0 | 6 | 5 | |
| GSE74703 | ACAP2 | 23527 | 19 | 0 | 17 | |
| GSE74704 | ACAP2 | 23527 | 14 | 0 | 6 | |
| TCGA | ACAP2 | 23527 | 77 | 1 | 18 |
Total number of gains: 265; Total number of losses: 32; Total Number of normals: 191.
Somatic mutations of ACAP2:
Generating mutation plots.
Highly correlated genes for ACAP2:
Showing top 20/514 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ACAP2 | PANK1 | 0.799546 | 3 | 0 | 3 |
| ACAP2 | PIK3C3 | 0.78391 | 3 | 0 | 3 |
| ACAP2 | PSMD1 | 0.772502 | 3 | 0 | 3 |
| ACAP2 | STAM2 | 0.758554 | 4 | 0 | 4 |
| ACAP2 | ZNF268 | 0.739687 | 3 | 0 | 3 |
| ACAP2 | MPPED2 | 0.738648 | 3 | 0 | 3 |
| ACAP2 | WARS2 | 0.736678 | 3 | 0 | 3 |
| ACAP2 | SLC33A1 | 0.734543 | 3 | 0 | 3 |
| ACAP2 | NCBP1 | 0.732863 | 3 | 0 | 3 |
| ACAP2 | POLE3 | 0.732605 | 3 | 0 | 3 |
| ACAP2 | PSMA5 | 0.730582 | 3 | 0 | 3 |
| ACAP2 | ARPC1A | 0.729921 | 3 | 0 | 3 |
| ACAP2 | GPN1 | 0.725896 | 3 | 0 | 3 |
| ACAP2 | XRN1 | 0.723019 | 3 | 0 | 3 |
| ACAP2 | GDAP2 | 0.717205 | 3 | 0 | 3 |
| ACAP2 | KCNK17 | 0.716172 | 3 | 0 | 3 |
| ACAP2 | RAB9A | 0.715476 | 5 | 0 | 5 |
| ACAP2 | AGGF1 | 0.708849 | 3 | 0 | 3 |
| ACAP2 | MAPKAP1 | 0.702587 | 3 | 0 | 3 |
| ACAP2 | ACSL1 | 0.701758 | 3 | 0 | 3 |
For details and further investigation, click here