| Full name: glycerol kinase 2 | Alias Symbol: GKTA | ||
| Type: protein-coding gene | Cytoband: 4q21.21 | ||
| Entrez ID: 2712 | HGNC ID: HGNC:4291 | Ensembl Gene: ENSG00000196475 | OMIM ID: 600148 |
| Drug and gene relationship at DGIdb | |||
GK2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa03320 | PPAR signaling pathway |
Expression of GK2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GK2 | 2712 | 215430_at | 0.1611 | 0.5377 | |
| GSE20347 | GK2 | 2712 | 215430_at | -0.0334 | 0.7502 | |
| GSE23400 | GK2 | 2712 | 215430_at | -0.1636 | 0.0003 | |
| GSE26886 | GK2 | 2712 | 215430_at | -0.1127 | 0.2391 | |
| GSE29001 | GK2 | 2712 | 215430_at | -0.1426 | 0.3659 | |
| GSE38129 | GK2 | 2712 | 215430_at | -0.0470 | 0.5941 | |
| GSE45670 | GK2 | 2712 | 215430_at | 0.1574 | 0.0901 | |
| GSE53622 | GK2 | 2712 | 11588 | 0.1087 | 0.3681 | |
| GSE53624 | GK2 | 2712 | 11588 | 0.0039 | 0.9657 | |
| GSE63941 | GK2 | 2712 | 215430_at | -0.0308 | 0.8875 | |
| GSE77861 | GK2 | 2712 | 215430_at | -0.0334 | 0.7725 |
Upregulated datasets: 0; Downregulated datasets: 0.
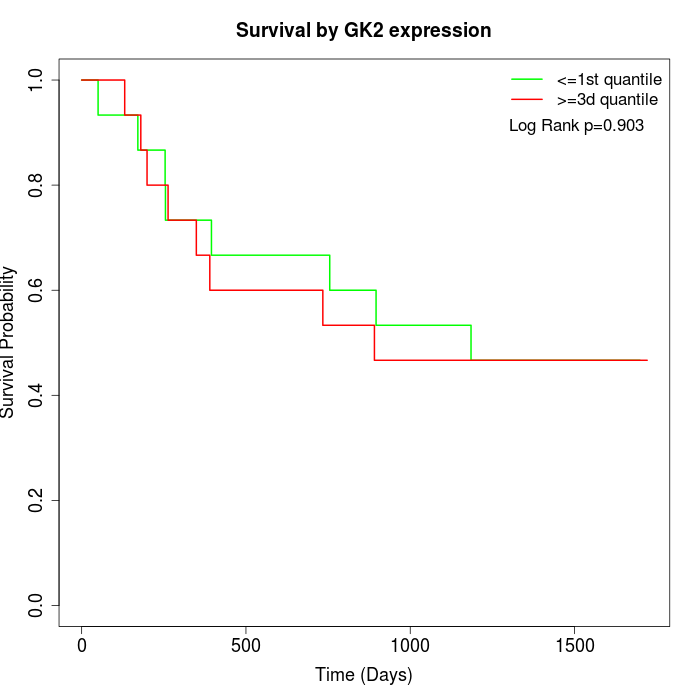
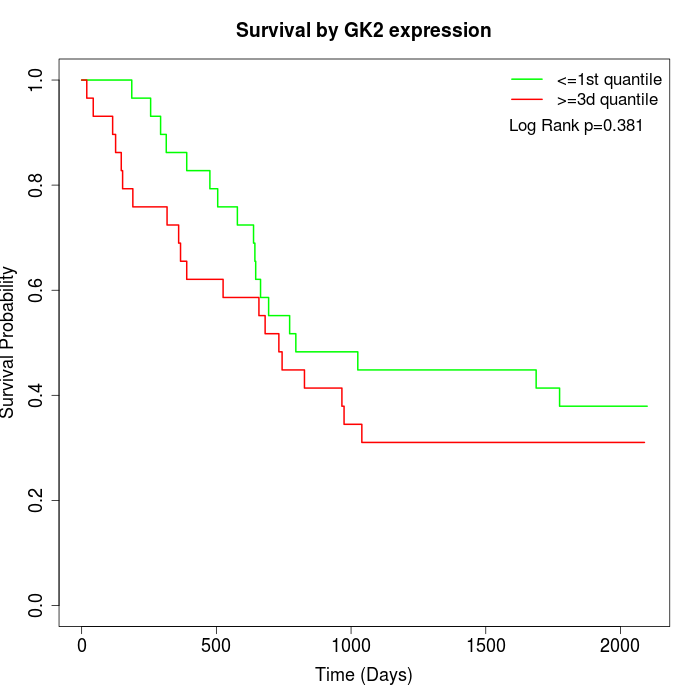
Survival by GK2 expression:
Note: Click image to view full size file.
Copy number change of GK2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GK2 | 2712 | 1 | 10 | 19 | |
| GSE20123 | GK2 | 2712 | 1 | 10 | 19 | |
| GSE43470 | GK2 | 2712 | 0 | 13 | 30 | |
| GSE46452 | GK2 | 2712 | 1 | 36 | 22 | |
| GSE47630 | GK2 | 2712 | 1 | 19 | 20 | |
| GSE54993 | GK2 | 2712 | 8 | 0 | 62 | |
| GSE54994 | GK2 | 2712 | 0 | 9 | 44 | |
| GSE60625 | GK2 | 2712 | 0 | 0 | 11 | |
| GSE74703 | GK2 | 2712 | 0 | 11 | 25 | |
| GSE74704 | GK2 | 2712 | 1 | 6 | 13 | |
| TCGA | GK2 | 2712 | 12 | 33 | 51 |
Total number of gains: 25; Total number of losses: 147; Total Number of normals: 316.
Somatic mutations of GK2:
Generating mutation plots.
Highly correlated genes for GK2:
Showing top 20/544 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GK2 | RALYL | 0.726162 | 3 | 0 | 3 |
| GK2 | MEFV | 0.726032 | 3 | 0 | 3 |
| GK2 | CGA | 0.724033 | 4 | 0 | 4 |
| GK2 | SERPINA4 | 0.710279 | 3 | 0 | 3 |
| GK2 | OR10J1 | 0.703364 | 4 | 0 | 4 |
| GK2 | IL9R | 0.702325 | 4 | 0 | 4 |
| GK2 | ZNF460 | 0.698219 | 4 | 0 | 4 |
| GK2 | NTNG1 | 0.695376 | 4 | 0 | 4 |
| GK2 | OR2J2 | 0.695009 | 3 | 0 | 3 |
| GK2 | SLC28A2 | 0.693939 | 5 | 0 | 5 |
| GK2 | HHLA1 | 0.692494 | 4 | 0 | 4 |
| GK2 | TSHR | 0.691715 | 3 | 0 | 3 |
| GK2 | SLC6A12 | 0.687217 | 4 | 0 | 4 |
| GK2 | GDF5 | 0.686586 | 4 | 0 | 4 |
| GK2 | SYT13 | 0.685966 | 3 | 0 | 3 |
| GK2 | DNTT | 0.685409 | 3 | 0 | 3 |
| GK2 | PSG4 | 0.68528 | 3 | 0 | 3 |
| GK2 | CCDC7 | 0.684526 | 4 | 0 | 4 |
| GK2 | KIR2DS2 | 0.683078 | 4 | 0 | 4 |
| GK2 | OR6A2 | 0.681912 | 4 | 0 | 4 |
For details and further investigation, click here