| Full name: glycoprotein hormone subunit alpha 2 | Alias Symbol: GPA2|ZSIG51|A2|MGC126572 | ||
| Type: protein-coding gene | Cytoband: 11q13.1 | ||
| Entrez ID: 170589 | HGNC ID: HGNC:18054 | Ensembl Gene: ENSG00000149735 | OMIM ID: 609651 |
| Drug and gene relationship at DGIdb | |||
Expression of GPHA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GPHA2 | 170589 | 237230_at | 0.0832 | 0.7809 | |
| GSE26886 | GPHA2 | 170589 | 237230_at | 0.0311 | 0.7470 | |
| GSE45670 | GPHA2 | 170589 | 237230_at | 0.1587 | 0.0691 | |
| GSE53622 | GPHA2 | 170589 | 103240 | 0.2481 | 0.0006 | |
| GSE53624 | GPHA2 | 170589 | 103240 | 0.1846 | 0.0059 | |
| GSE63941 | GPHA2 | 170589 | 237230_at | -0.0367 | 0.8190 | |
| GSE77861 | GPHA2 | 170589 | 237230_at | -0.0679 | 0.3436 | |
| GSE97050 | GPHA2 | 170589 | A_23_P47181 | 0.1098 | 0.5652 | |
| TCGA | GPHA2 | 170589 | RNAseq | -4.0293 | 0.0007 |
Upregulated datasets: 0; Downregulated datasets: 1.
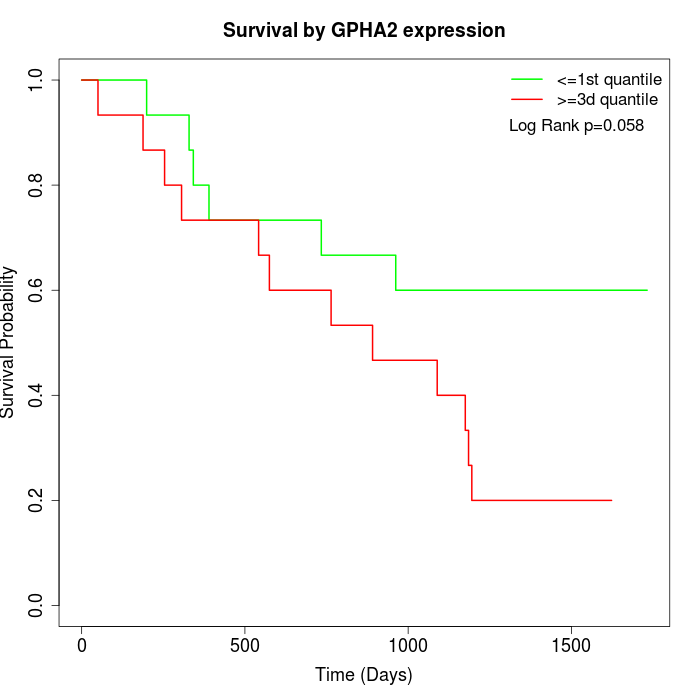
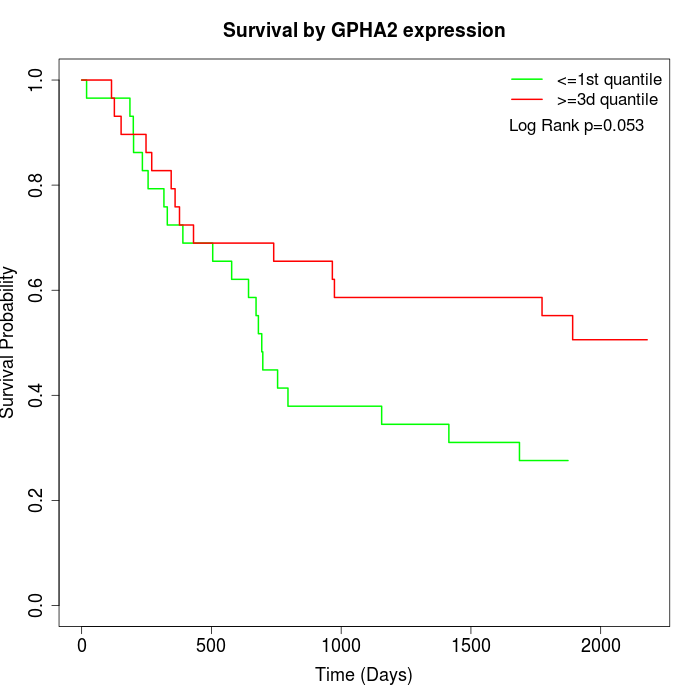
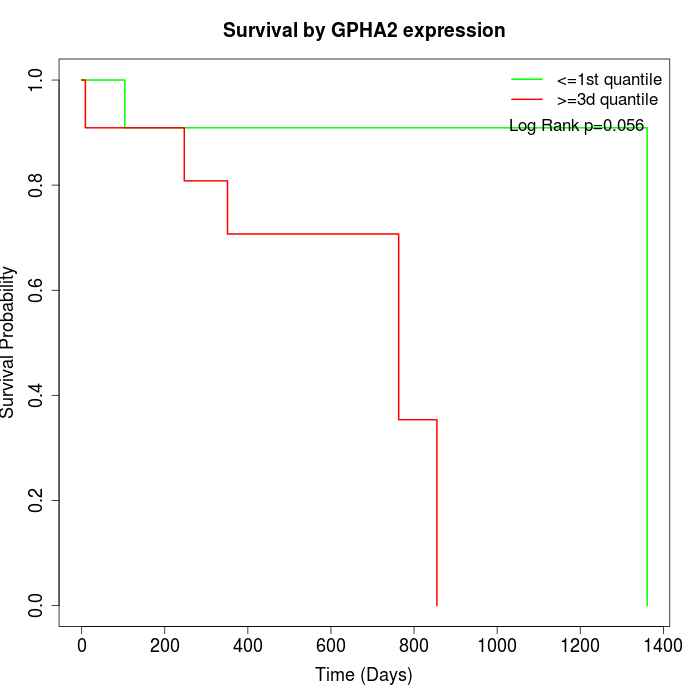
Survival by GPHA2 expression:
Note: Click image to view full size file.
Copy number change of GPHA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GPHA2 | 170589 | 7 | 6 | 17 | |
| GSE20123 | GPHA2 | 170589 | 7 | 6 | 17 | |
| GSE43470 | GPHA2 | 170589 | 1 | 3 | 39 | |
| GSE46452 | GPHA2 | 170589 | 10 | 4 | 45 | |
| GSE47630 | GPHA2 | 170589 | 5 | 6 | 29 | |
| GSE54993 | GPHA2 | 170589 | 3 | 0 | 67 | |
| GSE54994 | GPHA2 | 170589 | 6 | 5 | 42 | |
| GSE60625 | GPHA2 | 170589 | 0 | 3 | 8 | |
| GSE74703 | GPHA2 | 170589 | 1 | 1 | 34 | |
| GSE74704 | GPHA2 | 170589 | 5 | 4 | 11 | |
| TCGA | GPHA2 | 170589 | 23 | 7 | 66 |
Total number of gains: 68; Total number of losses: 45; Total Number of normals: 375.
Somatic mutations of GPHA2:
Generating mutation plots.
Highly correlated genes for GPHA2:
Showing top 20/274 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GPHA2 | BSX | 0.802016 | 3 | 0 | 3 |
| GPHA2 | OR5A1 | 0.788298 | 3 | 0 | 3 |
| GPHA2 | LCN9 | 0.779682 | 3 | 0 | 3 |
| GPHA2 | HECW1 | 0.770943 | 3 | 0 | 3 |
| GPHA2 | PHGR1 | 0.770706 | 3 | 0 | 3 |
| GPHA2 | MS4A18 | 0.763215 | 3 | 0 | 3 |
| GPHA2 | PABPN1L | 0.76287 | 3 | 0 | 3 |
| GPHA2 | C1QL2 | 0.762437 | 3 | 0 | 3 |
| GPHA2 | C2orf72 | 0.761207 | 4 | 0 | 4 |
| GPHA2 | PDE6C | 0.759556 | 3 | 0 | 3 |
| GPHA2 | PSMB11 | 0.759515 | 3 | 0 | 3 |
| GPHA2 | MAG | 0.759395 | 3 | 0 | 3 |
| GPHA2 | ODF3L2 | 0.755551 | 3 | 0 | 3 |
| GPHA2 | SYT1 | 0.753549 | 3 | 0 | 3 |
| GPHA2 | TEX13A | 0.74992 | 3 | 0 | 3 |
| GPHA2 | TBX21 | 0.747394 | 3 | 0 | 3 |
| GPHA2 | DCDC1 | 0.745702 | 3 | 0 | 3 |
| GPHA2 | EVX1 | 0.745665 | 3 | 0 | 3 |
| GPHA2 | CRYGC | 0.74555 | 3 | 0 | 3 |
| GPHA2 | OPN4 | 0.742607 | 3 | 0 | 3 |
For details and further investigation, click here