| Full name: proline, histidine and glycine rich 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 15q15.1 | ||
| Entrez ID: 644844 | HGNC ID: HGNC:37226 | Ensembl Gene: ENSG00000233041 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of PHGR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PHGR1 | 644844 | 230485_at | 0.0313 | 0.9177 | |
| GSE26886 | PHGR1 | 644844 | 230485_at | 0.2524 | 0.0344 | |
| GSE45670 | PHGR1 | 644844 | 230485_at | 0.1136 | 0.1356 | |
| GSE53622 | PHGR1 | 644844 | 40636 | 0.1592 | 0.5239 | |
| GSE53624 | PHGR1 | 644844 | 40636 | -0.4839 | 0.0109 | |
| GSE63941 | PHGR1 | 644844 | 230485_at | 0.4258 | 0.0048 | |
| GSE77861 | PHGR1 | 644844 | 230485_at | -0.1235 | 0.4938 | |
| GSE97050 | PHGR1 | 644844 | A_33_P3323501 | -0.0207 | 0.9287 | |
| TCGA | PHGR1 | 644844 | RNAseq | -6.1476 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
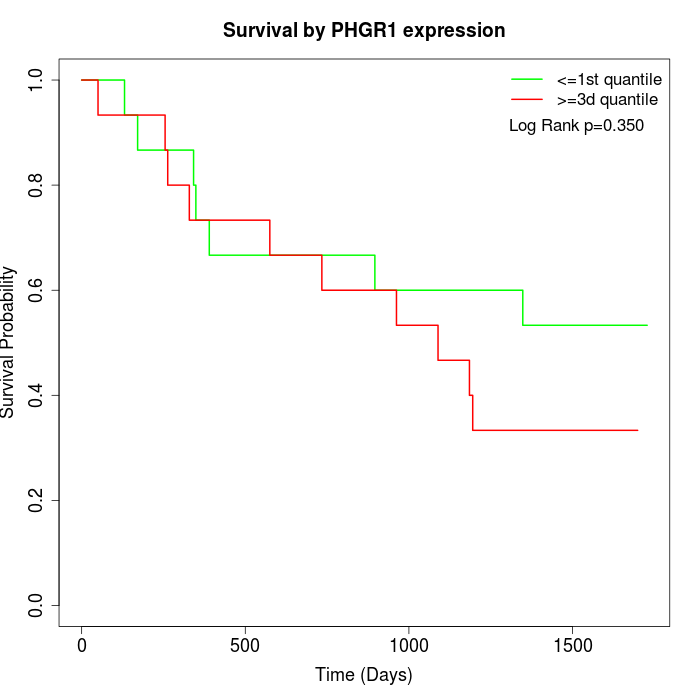
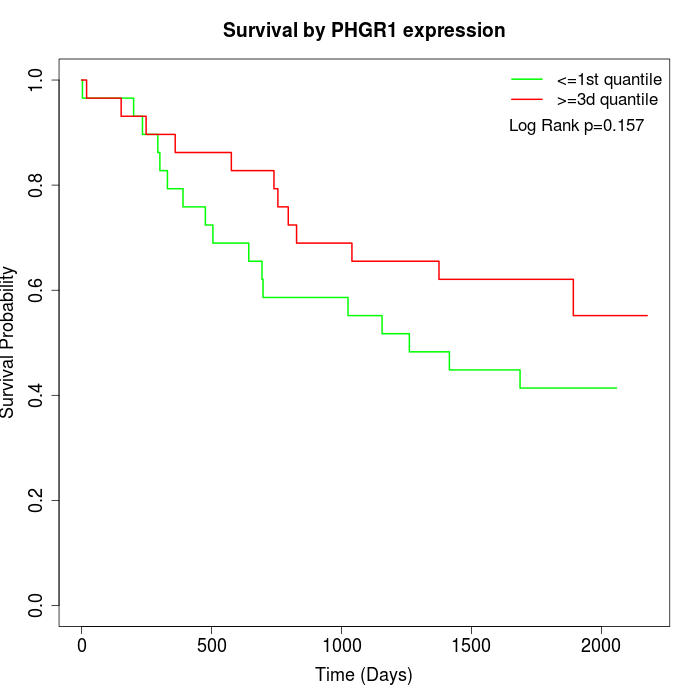
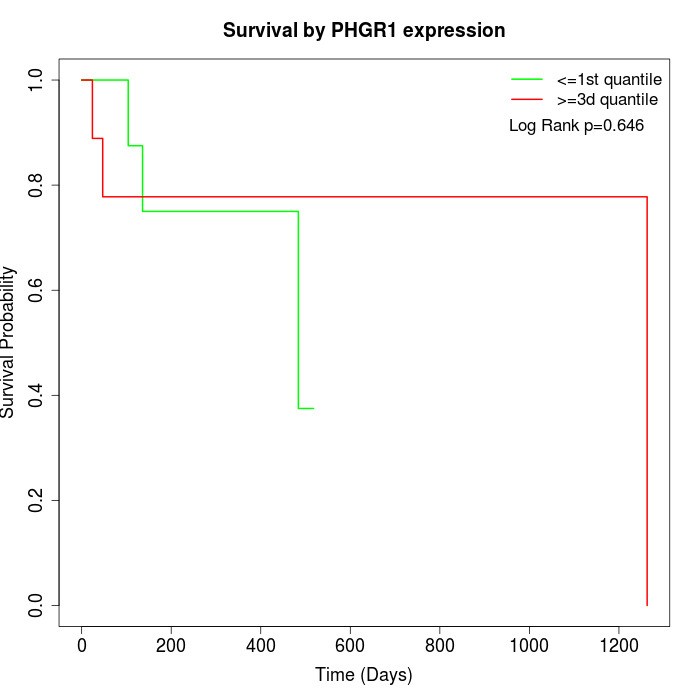
Survival by PHGR1 expression:
Note: Click image to view full size file.
Copy number change of PHGR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PHGR1 | 644844 | 4 | 5 | 21 | |
| GSE20123 | PHGR1 | 644844 | 4 | 5 | 21 | |
| GSE43470 | PHGR1 | 644844 | 4 | 6 | 33 | |
| GSE46452 | PHGR1 | 644844 | 3 | 7 | 49 | |
| GSE47630 | PHGR1 | 644844 | 8 | 11 | 21 | |
| GSE54993 | PHGR1 | 644844 | 4 | 6 | 60 | |
| GSE54994 | PHGR1 | 644844 | 5 | 7 | 41 | |
| GSE60625 | PHGR1 | 644844 | 4 | 0 | 7 | |
| GSE74703 | PHGR1 | 644844 | 4 | 5 | 27 | |
| GSE74704 | PHGR1 | 644844 | 3 | 3 | 14 | |
| TCGA | PHGR1 | 644844 | 11 | 17 | 68 |
Total number of gains: 54; Total number of losses: 72; Total Number of normals: 362.
Somatic mutations of PHGR1:
Generating mutation plots.
Highly correlated genes for PHGR1:
Showing top 20/281 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PHGR1 | CTSE | 0.892731 | 3 | 0 | 3 |
| PHGR1 | PGC | 0.859643 | 4 | 0 | 4 |
| PHGR1 | ATP2B2 | 0.777566 | 3 | 0 | 3 |
| PHGR1 | GPHA2 | 0.770706 | 3 | 0 | 3 |
| PHGR1 | NOX1 | 0.758515 | 3 | 0 | 3 |
| PHGR1 | SCN2B | 0.74384 | 3 | 0 | 3 |
| PHGR1 | PHOSPHO1 | 0.741414 | 3 | 0 | 3 |
| PHGR1 | ZIC1 | 0.739118 | 3 | 0 | 3 |
| PHGR1 | PPP2R5D | 0.739025 | 3 | 0 | 3 |
| PHGR1 | UMODL1 | 0.738692 | 3 | 0 | 3 |
| PHGR1 | RCOR2 | 0.737146 | 3 | 0 | 3 |
| PHGR1 | DUOXA2 | 0.735913 | 3 | 0 | 3 |
| PHGR1 | ISLR2 | 0.734333 | 3 | 0 | 3 |
| PHGR1 | JSRP1 | 0.733612 | 3 | 0 | 3 |
| PHGR1 | C11orf21 | 0.732735 | 3 | 0 | 3 |
| PHGR1 | PLA2G4F | 0.732457 | 3 | 0 | 3 |
| PHGR1 | MEX3D | 0.731563 | 3 | 0 | 3 |
| PHGR1 | RAX | 0.731225 | 3 | 0 | 3 |
| PHGR1 | DNAH6 | 0.730903 | 3 | 0 | 3 |
| PHGR1 | PNPLA2 | 0.73029 | 3 | 0 | 3 |
For details and further investigation, click here