| Full name: glutathione peroxidase 5 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 6p22.1 | ||
| Entrez ID: 2880 | HGNC ID: HGNC:4557 | Ensembl Gene: ENSG00000224586 | OMIM ID: 603435 |
| Drug and gene relationship at DGIdb | |||
Expression of GPX5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GPX5 | 2880 | 214648_at | -0.1639 | 0.5034 | |
| GSE20347 | GPX5 | 2880 | 214648_at | -0.1548 | 0.0124 | |
| GSE23400 | GPX5 | 2880 | 208028_s_at | -0.1757 | 0.0001 | |
| GSE26886 | GPX5 | 2880 | 214648_at | -0.1926 | 0.0540 | |
| GSE29001 | GPX5 | 2880 | 214648_at | -0.2052 | 0.0989 | |
| GSE38129 | GPX5 | 2880 | 214648_at | -0.2495 | 0.0000 | |
| GSE45670 | GPX5 | 2880 | 208028_s_at | 0.0750 | 0.3594 | |
| GSE53622 | GPX5 | 2880 | 33949 | 0.4849 | 0.0000 | |
| GSE53624 | GPX5 | 2880 | 33949 | 0.0793 | 0.6550 | |
| GSE63941 | GPX5 | 2880 | 208028_s_at | 0.0239 | 0.8588 | |
| GSE77861 | GPX5 | 2880 | 208028_s_at | -0.0103 | 0.9350 | |
| TCGA | GPX5 | 2880 | RNAseq | 1.2164 | 0.5412 |
Upregulated datasets: 0; Downregulated datasets: 0.
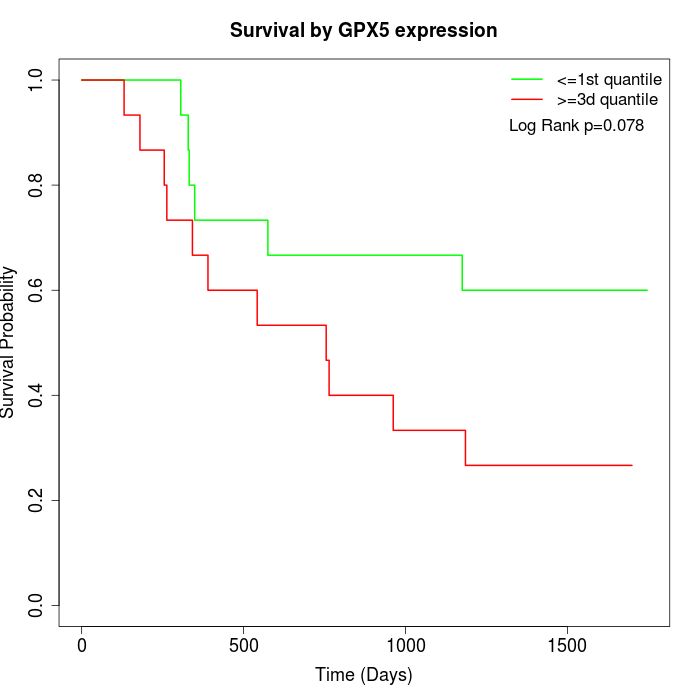
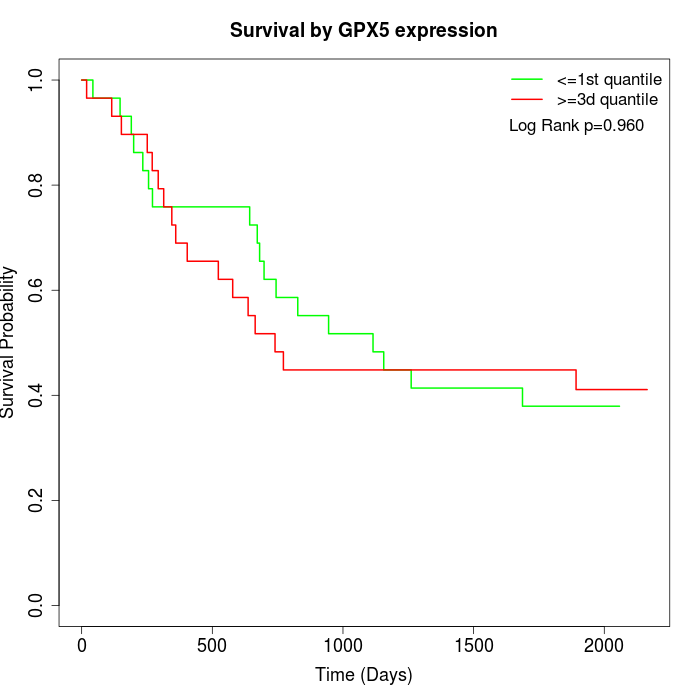
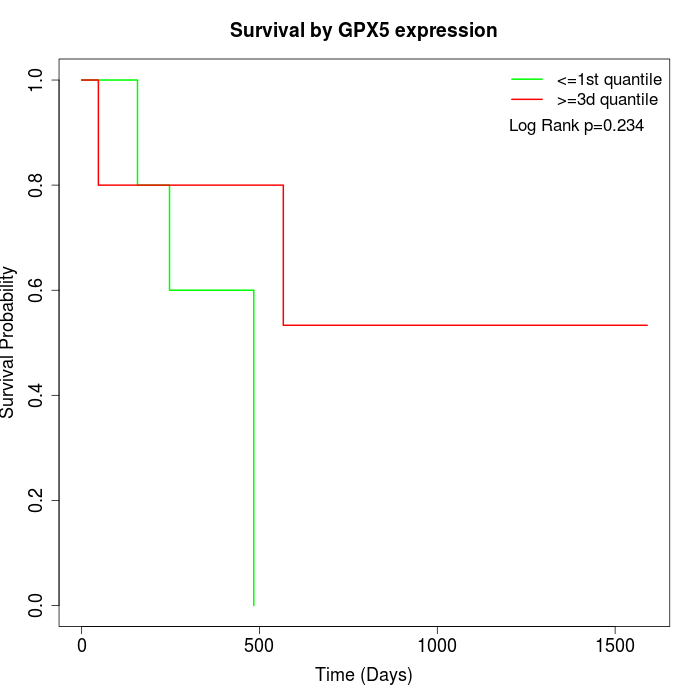
Survival by GPX5 expression:
Note: Click image to view full size file.
Copy number change of GPX5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GPX5 | 2880 | 3 | 2 | 25 | |
| GSE20123 | GPX5 | 2880 | 3 | 2 | 25 | |
| GSE43470 | GPX5 | 2880 | 5 | 0 | 38 | |
| GSE46452 | GPX5 | 2880 | 1 | 10 | 48 | |
| GSE47630 | GPX5 | 2880 | 7 | 4 | 29 | |
| GSE54993 | GPX5 | 2880 | 2 | 1 | 67 | |
| GSE54994 | GPX5 | 2880 | 11 | 4 | 38 | |
| GSE60625 | GPX5 | 2880 | 0 | 1 | 10 | |
| GSE74703 | GPX5 | 2880 | 5 | 0 | 31 | |
| GSE74704 | GPX5 | 2880 | 0 | 1 | 19 | |
| TCGA | GPX5 | 2880 | 16 | 19 | 61 |
Total number of gains: 53; Total number of losses: 44; Total Number of normals: 391.
Somatic mutations of GPX5:
Generating mutation plots.
Highly correlated genes for GPX5:
Showing top 20/423 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GPX5 | PNPLA7 | 0.706103 | 3 | 0 | 3 |
| GPX5 | SLC25A53 | 0.693997 | 3 | 0 | 3 |
| GPX5 | LINC00917 | 0.682065 | 3 | 0 | 3 |
| GPX5 | CGA | 0.680541 | 3 | 0 | 3 |
| GPX5 | CER1 | 0.67887 | 3 | 0 | 3 |
| GPX5 | RASGRP2 | 0.678748 | 3 | 0 | 3 |
| GPX5 | PHLPP2 | 0.677236 | 3 | 0 | 3 |
| GPX5 | ZNF248 | 0.676192 | 3 | 0 | 3 |
| GPX5 | LINC01350 | 0.67549 | 3 | 0 | 3 |
| GPX5 | AKAP6 | 0.668393 | 3 | 0 | 3 |
| GPX5 | TAAR2 | 0.660673 | 4 | 0 | 3 |
| GPX5 | PSG9 | 0.658156 | 3 | 0 | 3 |
| GPX5 | WDR87 | 0.646223 | 3 | 0 | 3 |
| GPX5 | PRODH2 | 0.643663 | 5 | 0 | 5 |
| GPX5 | LINC00276 | 0.641719 | 3 | 0 | 3 |
| GPX5 | CAMKV | 0.637883 | 5 | 0 | 5 |
| GPX5 | ASCL3 | 0.636827 | 4 | 0 | 3 |
| GPX5 | LINC01282 | 0.634693 | 3 | 0 | 3 |
| GPX5 | GRM1 | 0.634216 | 4 | 0 | 4 |
| GPX5 | EPS8L3 | 0.630573 | 5 | 0 | 4 |
For details and further investigation, click here