| Full name: cerberus 1, DAN family BMP antagonist | Alias Symbol: DAND4 | ||
| Type: protein-coding gene | Cytoband: 9p22.3 | ||
| Entrez ID: 9350 | HGNC ID: HGNC:1862 | Ensembl Gene: ENSG00000147869 | OMIM ID: 603777 |
| Drug and gene relationship at DGIdb | |||
CER1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04310 | Wnt signaling pathway |
Expression of CER1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CER1 | 9350 | 221378_at | -0.1197 | 0.5707 | |
| GSE20347 | CER1 | 9350 | 221378_at | 0.0111 | 0.9224 | |
| GSE23400 | CER1 | 9350 | 221378_at | -0.2062 | 0.0001 | |
| GSE26886 | CER1 | 9350 | 221378_at | -0.0264 | 0.8556 | |
| GSE29001 | CER1 | 9350 | 221378_at | -0.2013 | 0.2062 | |
| GSE38129 | CER1 | 9350 | 221378_at | -0.1830 | 0.0603 | |
| GSE45670 | CER1 | 9350 | 221378_at | 0.0423 | 0.7053 | |
| GSE53622 | CER1 | 9350 | 81213 | 0.3263 | 0.0405 | |
| GSE53624 | CER1 | 9350 | 81213 | 0.5291 | 0.0001 | |
| GSE63941 | CER1 | 9350 | 221378_at | 0.4771 | 0.0016 | |
| GSE77861 | CER1 | 9350 | 221378_at | -0.1620 | 0.2276 |
Upregulated datasets: 0; Downregulated datasets: 0.
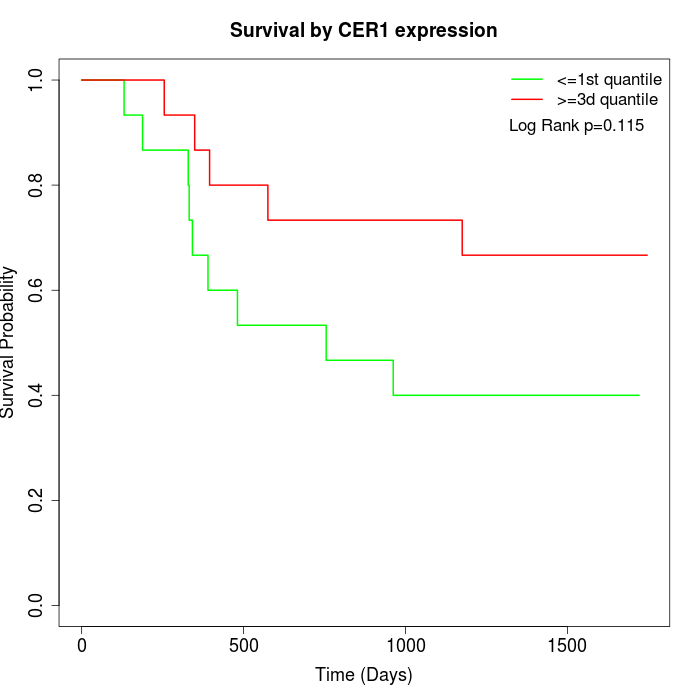
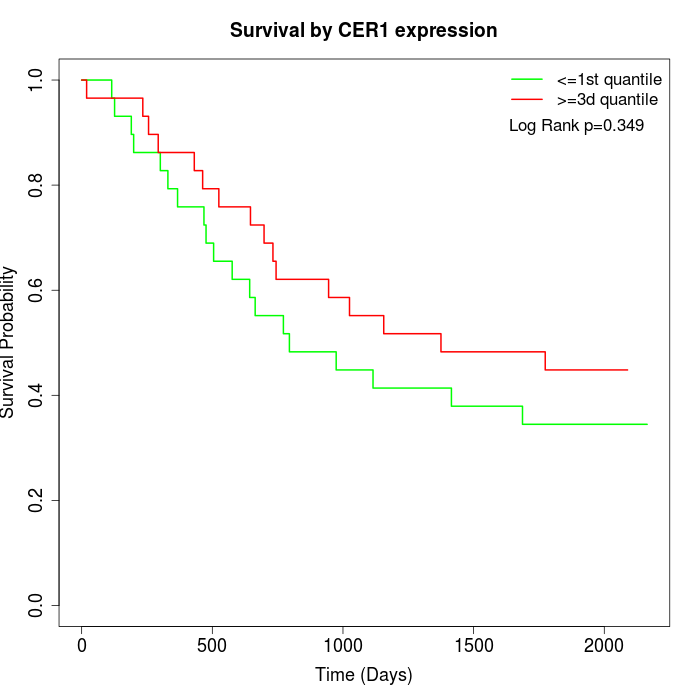
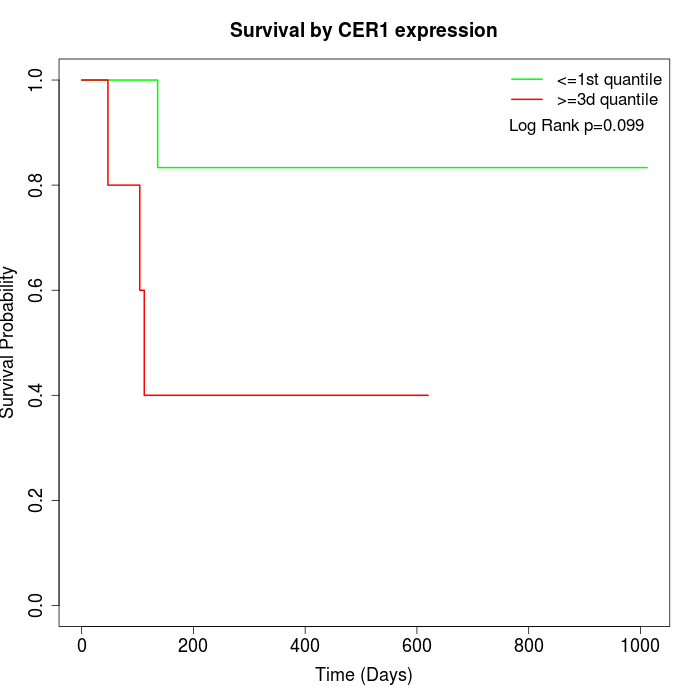
Survival by CER1 expression:
Note: Click image to view full size file.
Copy number change of CER1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CER1 | 9350 | 2 | 16 | 12 | |
| GSE20123 | CER1 | 9350 | 2 | 16 | 12 | |
| GSE43470 | CER1 | 9350 | 5 | 11 | 27 | |
| GSE46452 | CER1 | 9350 | 2 | 23 | 34 | |
| GSE47630 | CER1 | 9350 | 4 | 26 | 10 | |
| GSE54993 | CER1 | 9350 | 6 | 3 | 61 | |
| GSE54994 | CER1 | 9350 | 10 | 16 | 27 | |
| GSE60625 | CER1 | 9350 | 0 | 0 | 11 | |
| GSE74703 | CER1 | 9350 | 5 | 8 | 23 | |
| GSE74704 | CER1 | 9350 | 0 | 13 | 7 | |
| TCGA | CER1 | 9350 | 14 | 50 | 32 |
Total number of gains: 50; Total number of losses: 182; Total Number of normals: 256.
Somatic mutations of CER1:
Generating mutation plots.
Highly correlated genes for CER1:
Showing top 20/1165 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CER1 | FSTL4 | 0.762144 | 4 | 0 | 4 |
| CER1 | MTAP | 0.758505 | 5 | 0 | 5 |
| CER1 | UBE2D4 | 0.753371 | 3 | 0 | 3 |
| CER1 | GAD2 | 0.752912 | 4 | 0 | 4 |
| CER1 | NMNAT2 | 0.748754 | 3 | 0 | 3 |
| CER1 | AQP6 | 0.747872 | 4 | 0 | 4 |
| CER1 | PHF7 | 0.741267 | 4 | 0 | 4 |
| CER1 | FOLH1 | 0.738296 | 4 | 0 | 4 |
| CER1 | BRINP2 | 0.737554 | 5 | 0 | 5 |
| CER1 | AKAP6 | 0.735535 | 4 | 0 | 4 |
| CER1 | OTOR | 0.730133 | 4 | 0 | 4 |
| CER1 | SLC22A13 | 0.727006 | 4 | 0 | 4 |
| CER1 | ARPP21 | 0.725794 | 4 | 0 | 4 |
| CER1 | KRT38 | 0.723002 | 4 | 0 | 4 |
| CER1 | SYT13 | 0.717337 | 4 | 0 | 3 |
| CER1 | GDF2 | 0.717052 | 4 | 0 | 4 |
| CER1 | FSCN3 | 0.716719 | 6 | 0 | 6 |
| CER1 | GPR12 | 0.715846 | 4 | 0 | 3 |
| CER1 | NTNG1 | 0.714704 | 4 | 0 | 4 |
| CER1 | FRAS1 | 0.714606 | 5 | 0 | 5 |
For details and further investigation, click here