| Full name: G1 to S phase transition 2 | Alias Symbol: eRF3b|FLJ10441 | ||
| Type: protein-coding gene | Cytoband: Xp11.22 | ||
| Entrez ID: 23708 | HGNC ID: HGNC:4622 | Ensembl Gene: ENSG00000189369 | OMIM ID: 300418 |
| Drug and gene relationship at DGIdb | |||
Expression of GSPT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GSPT2 | 23708 | 205541_s_at | -0.1679 | 0.9088 | |
| GSE20347 | GSPT2 | 23708 | 205541_s_at | 0.2253 | 0.5285 | |
| GSE23400 | GSPT2 | 23708 | 205541_s_at | 0.0564 | 0.5635 | |
| GSE26886 | GSPT2 | 23708 | 205541_s_at | 0.8199 | 0.1565 | |
| GSE29001 | GSPT2 | 23708 | 205541_s_at | -0.1556 | 0.7927 | |
| GSE38129 | GSPT2 | 23708 | 205541_s_at | 0.1258 | 0.7323 | |
| GSE45670 | GSPT2 | 23708 | 205541_s_at | -0.1725 | 0.7566 | |
| GSE53622 | GSPT2 | 23708 | 110051 | 0.2006 | 0.0162 | |
| GSE53624 | GSPT2 | 23708 | 110051 | 0.3343 | 0.0000 | |
| GSE63941 | GSPT2 | 23708 | 205541_s_at | -4.0001 | 0.0389 | |
| GSE77861 | GSPT2 | 23708 | 205541_s_at | 0.7326 | 0.1776 | |
| GSE97050 | GSPT2 | 23708 | A_23_P19115 | 0.2723 | 0.3828 | |
| SRP007169 | GSPT2 | 23708 | RNAseq | 0.1444 | 0.8317 | |
| SRP064894 | GSPT2 | 23708 | RNAseq | 0.3511 | 0.1597 | |
| SRP133303 | GSPT2 | 23708 | RNAseq | 0.6220 | 0.1241 | |
| SRP159526 | GSPT2 | 23708 | RNAseq | 0.9569 | 0.0748 | |
| SRP193095 | GSPT2 | 23708 | RNAseq | 0.4299 | 0.0465 | |
| SRP219564 | GSPT2 | 23708 | RNAseq | -0.1631 | 0.7581 | |
| TCGA | GSPT2 | 23708 | RNAseq | -0.3501 | 0.0780 |
Upregulated datasets: 0; Downregulated datasets: 1.
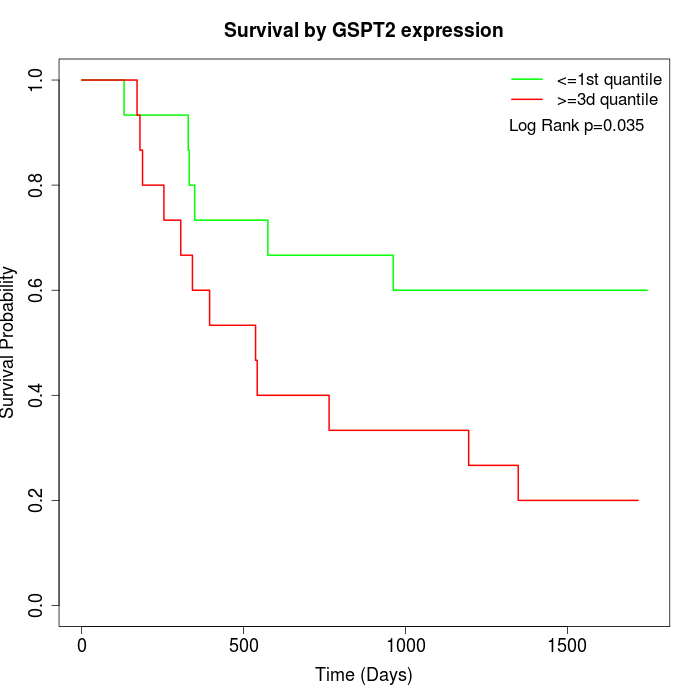
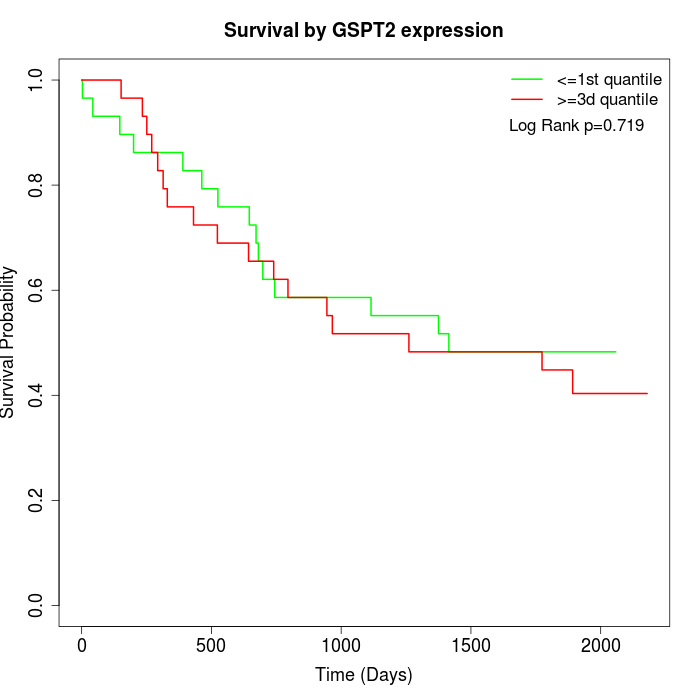
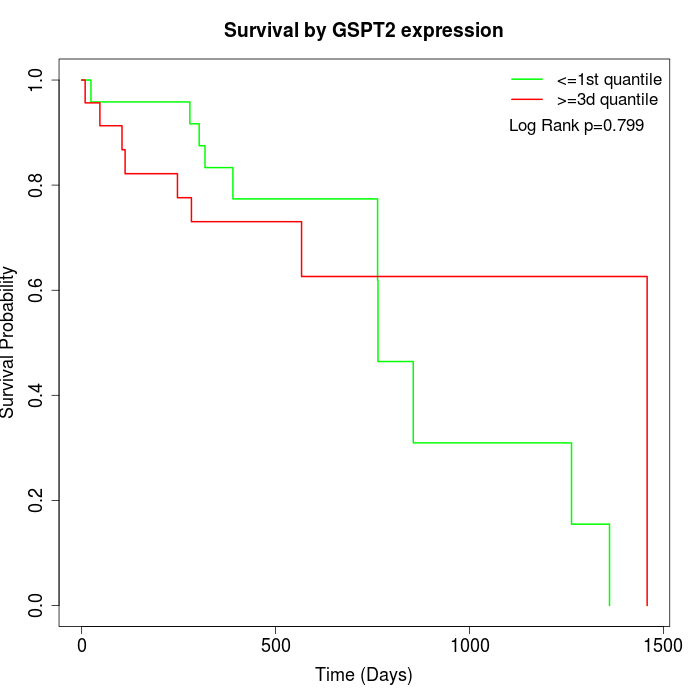
Survival by GSPT2 expression:
Note: Click image to view full size file.
Copy number change of GSPT2:
No record found for this gene.
Somatic mutations of GSPT2:
Generating mutation plots.
Highly correlated genes for GSPT2:
Showing top 20/388 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GSPT2 | ZNF616 | 0.796962 | 3 | 0 | 3 |
| GSPT2 | UBQLN2 | 0.767512 | 3 | 0 | 3 |
| GSPT2 | POLR2L | 0.758669 | 3 | 0 | 3 |
| GSPT2 | SRSF10 | 0.750677 | 3 | 0 | 3 |
| GSPT2 | ZNF260 | 0.747412 | 3 | 0 | 3 |
| GSPT2 | DEFB132 | 0.744335 | 3 | 0 | 3 |
| GSPT2 | THAP5 | 0.743355 | 3 | 0 | 3 |
| GSPT2 | CREB3 | 0.741603 | 3 | 0 | 3 |
| GSPT2 | ZNF546 | 0.73874 | 3 | 0 | 3 |
| GSPT2 | ZNF398 | 0.736044 | 3 | 0 | 3 |
| GSPT2 | PTPN1 | 0.732135 | 3 | 0 | 3 |
| GSPT2 | ZNF10 | 0.728535 | 4 | 0 | 4 |
| GSPT2 | FKBP7 | 0.721587 | 3 | 0 | 3 |
| GSPT2 | AGBL5 | 0.72114 | 3 | 0 | 3 |
| GSPT2 | FAM171B | 0.717283 | 4 | 0 | 4 |
| GSPT2 | CWF19L2 | 0.717049 | 3 | 0 | 3 |
| GSPT2 | MNS1 | 0.716592 | 3 | 0 | 3 |
| GSPT2 | ZNF506 | 0.714965 | 3 | 0 | 3 |
| GSPT2 | MPG | 0.712844 | 3 | 0 | 3 |
| GSPT2 | FIBCD1 | 0.71268 | 3 | 0 | 3 |
For details and further investigation, click here