| Full name: potassium calcium-activated channel subfamily N member 4 | Alias Symbol: KCa3.1|hSK4|hKCa4|hIKCa1|IK | ||
| Type: protein-coding gene | Cytoband: 19q13.31 | ||
| Entrez ID: 3783 | HGNC ID: HGNC:6293 | Ensembl Gene: ENSG00000104783 | OMIM ID: 602754 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
KCNN4 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04970 | Salivary secretion |
Expression of KCNN4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNN4 | 3783 | 204401_at | -0.0334 | 0.9518 | |
| GSE20347 | KCNN4 | 3783 | 204401_at | 0.3906 | 0.1452 | |
| GSE23400 | KCNN4 | 3783 | 204401_at | -0.1104 | 0.1311 | |
| GSE26886 | KCNN4 | 3783 | 204401_at | 0.5946 | 0.0344 | |
| GSE29001 | KCNN4 | 3783 | 204401_at | 0.4215 | 0.4276 | |
| GSE38129 | KCNN4 | 3783 | 204401_at | 0.3316 | 0.2223 | |
| GSE45670 | KCNN4 | 3783 | 204401_at | 0.0178 | 0.9496 | |
| GSE53622 | KCNN4 | 3783 | 95534 | -0.0159 | 0.9400 | |
| GSE53624 | KCNN4 | 3783 | 95534 | -0.1314 | 0.3844 | |
| GSE63941 | KCNN4 | 3783 | 204401_at | 1.3817 | 0.0979 | |
| GSE77861 | KCNN4 | 3783 | 204401_at | 0.2844 | 0.5212 | |
| GSE97050 | KCNN4 | 3783 | A_23_P67529 | 0.2109 | 0.4586 | |
| SRP007169 | KCNN4 | 3783 | RNAseq | 2.3529 | 0.0084 | |
| SRP064894 | KCNN4 | 3783 | RNAseq | -0.0578 | 0.9063 | |
| SRP133303 | KCNN4 | 3783 | RNAseq | -0.0607 | 0.8940 | |
| SRP193095 | KCNN4 | 3783 | RNAseq | 0.3467 | 0.4859 | |
| SRP219564 | KCNN4 | 3783 | RNAseq | 0.0272 | 0.9687 | |
| TCGA | KCNN4 | 3783 | RNAseq | -0.3007 | 0.1378 |
Upregulated datasets: 1; Downregulated datasets: 0.
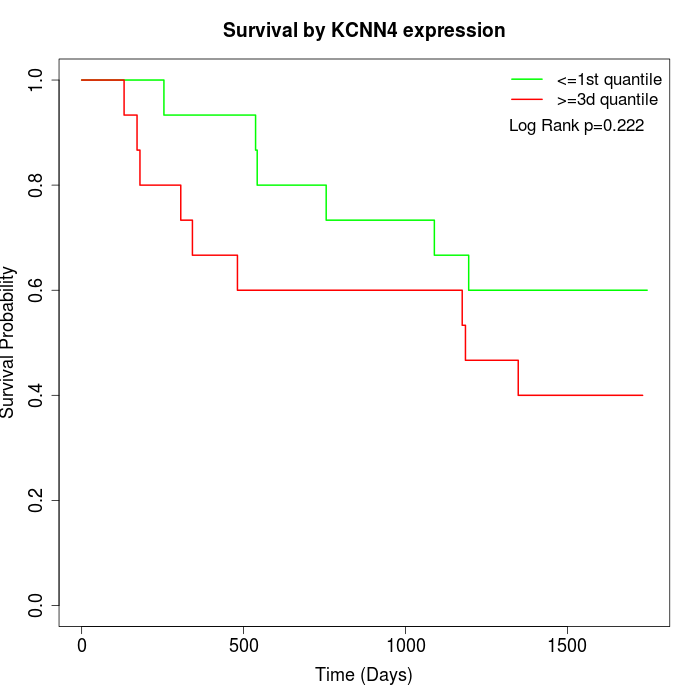
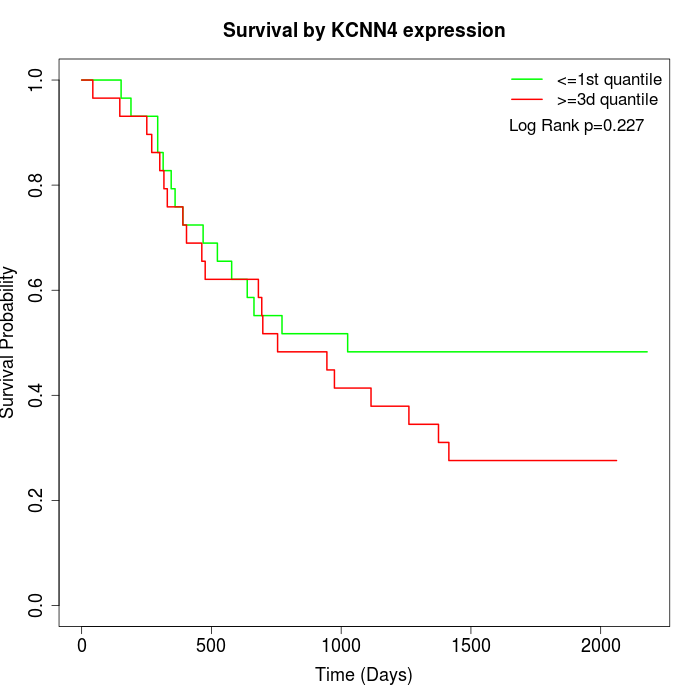
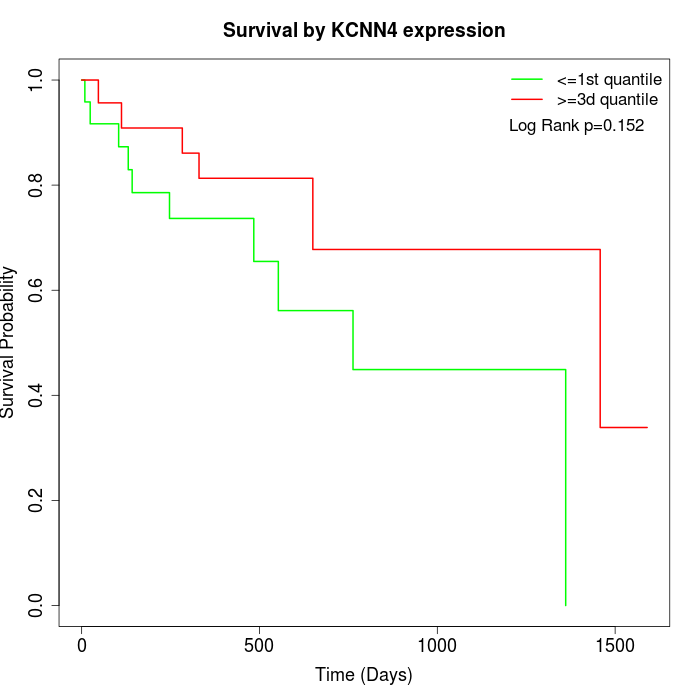
Survival by KCNN4 expression:
Note: Click image to view full size file.
Copy number change of KCNN4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNN4 | 3783 | 6 | 5 | 19 | |
| GSE20123 | KCNN4 | 3783 | 5 | 4 | 21 | |
| GSE43470 | KCNN4 | 3783 | 4 | 11 | 28 | |
| GSE46452 | KCNN4 | 3783 | 45 | 1 | 13 | |
| GSE47630 | KCNN4 | 3783 | 8 | 6 | 26 | |
| GSE54993 | KCNN4 | 3783 | 17 | 4 | 49 | |
| GSE54994 | KCNN4 | 3783 | 6 | 12 | 35 | |
| GSE60625 | KCNN4 | 3783 | 9 | 0 | 2 | |
| GSE74703 | KCNN4 | 3783 | 4 | 7 | 25 | |
| GSE74704 | KCNN4 | 3783 | 4 | 2 | 14 | |
| TCGA | KCNN4 | 3783 | 14 | 16 | 66 |
Total number of gains: 122; Total number of losses: 68; Total Number of normals: 298.
Somatic mutations of KCNN4:
Generating mutation plots.
Highly correlated genes for KCNN4:
Showing top 20/108 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNN4 | IL12RB2 | 0.777447 | 3 | 0 | 3 |
| KCNN4 | ZC3H10 | 0.747553 | 3 | 0 | 3 |
| KCNN4 | C1QTNF6 | 0.714723 | 3 | 0 | 3 |
| KCNN4 | TRIM10 | 0.712086 | 3 | 0 | 3 |
| KCNN4 | SLC29A2 | 0.691679 | 4 | 0 | 4 |
| KCNN4 | SALL4 | 0.681732 | 3 | 0 | 3 |
| KCNN4 | TEKT2 | 0.673572 | 4 | 0 | 3 |
| KCNN4 | RHBDD3 | 0.671634 | 3 | 0 | 3 |
| KCNN4 | ADD2 | 0.66398 | 4 | 0 | 3 |
| KCNN4 | TMEM63A | 0.657909 | 4 | 0 | 3 |
| KCNN4 | C6orf15 | 0.652372 | 3 | 0 | 3 |
| KCNN4 | DNAJC5B | 0.644772 | 3 | 0 | 3 |
| KCNN4 | NINJ2 | 0.644271 | 3 | 0 | 3 |
| KCNN4 | LYL1 | 0.640792 | 4 | 0 | 4 |
| KCNN4 | CSH1 | 0.639218 | 3 | 0 | 3 |
| KCNN4 | TMEM102 | 0.638875 | 3 | 0 | 3 |
| KCNN4 | PLB1 | 0.637867 | 3 | 0 | 3 |
| KCNN4 | GRM4 | 0.635252 | 4 | 0 | 4 |
| KCNN4 | VSIG4 | 0.632937 | 3 | 0 | 3 |
| KCNN4 | POLG2 | 0.632304 | 3 | 0 | 3 |
For details and further investigation, click here