| Full name: potassium channel tetramerization domain containing 3 | Alias Symbol: NY-REN-45 | ||
| Type: protein-coding gene | Cytoband: 1q41 | ||
| Entrez ID: 51133 | HGNC ID: HGNC:21305 | Ensembl Gene: ENSG00000136636 | OMIM ID: 613272 |
| Drug and gene relationship at DGIdb | |||
Expression of KCTD3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCTD3 | 51133 | 217894_at | -0.6054 | 0.1985 | |
| GSE20347 | KCTD3 | 51133 | 217894_at | -0.8795 | 0.0000 | |
| GSE23400 | KCTD3 | 51133 | 217894_at | -0.5866 | 0.0000 | |
| GSE26886 | KCTD3 | 51133 | 217894_at | -1.5599 | 0.0003 | |
| GSE29001 | KCTD3 | 51133 | 217894_at | -0.3944 | 0.1804 | |
| GSE38129 | KCTD3 | 51133 | 217894_at | -0.5783 | 0.0007 | |
| GSE45670 | KCTD3 | 51133 | 217894_at | -0.7236 | 0.0005 | |
| GSE53622 | KCTD3 | 51133 | 51795 | -0.8621 | 0.0000 | |
| GSE53624 | KCTD3 | 51133 | 51795 | -0.9941 | 0.0000 | |
| GSE63941 | KCTD3 | 51133 | 217894_at | -0.7883 | 0.1249 | |
| GSE77861 | KCTD3 | 51133 | 217894_at | -0.9382 | 0.0100 | |
| GSE97050 | KCTD3 | 51133 | A_23_P160406 | -0.4467 | 0.1421 | |
| SRP007169 | KCTD3 | 51133 | RNAseq | -1.8826 | 0.0000 | |
| SRP008496 | KCTD3 | 51133 | RNAseq | -1.6456 | 0.0000 | |
| SRP064894 | KCTD3 | 51133 | RNAseq | -1.2123 | 0.0000 | |
| SRP133303 | KCTD3 | 51133 | RNAseq | -0.7399 | 0.0003 | |
| SRP159526 | KCTD3 | 51133 | RNAseq | -0.6737 | 0.0666 | |
| SRP193095 | KCTD3 | 51133 | RNAseq | -0.9664 | 0.0000 | |
| SRP219564 | KCTD3 | 51133 | RNAseq | -0.8878 | 0.0101 | |
| TCGA | KCTD3 | 51133 | RNAseq | -0.1795 | 0.0005 |
Upregulated datasets: 0; Downregulated datasets: 4.
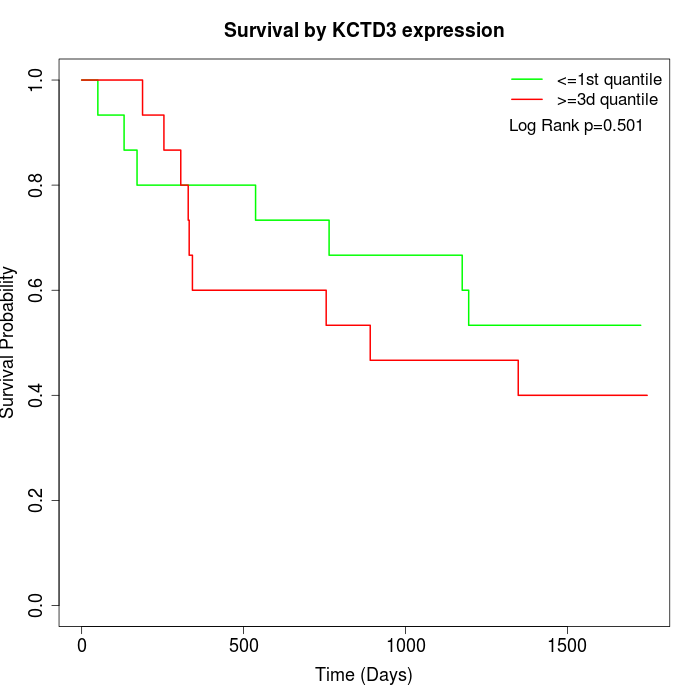
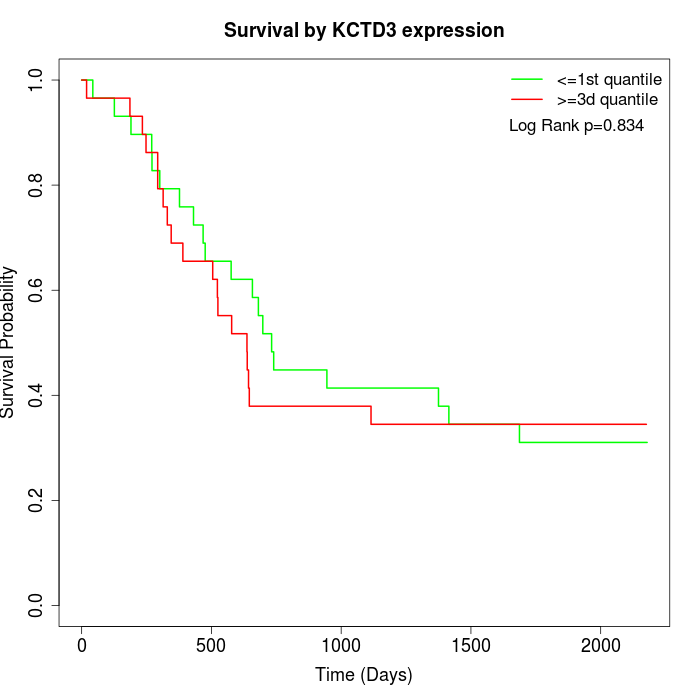
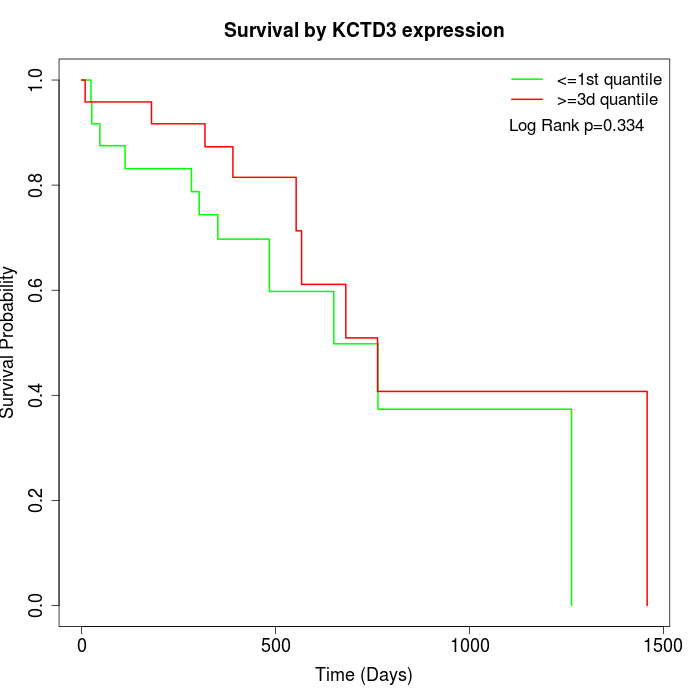
Survival by KCTD3 expression:
Note: Click image to view full size file.
Copy number change of KCTD3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCTD3 | 51133 | 9 | 0 | 21 | |
| GSE20123 | KCTD3 | 51133 | 9 | 0 | 21 | |
| GSE43470 | KCTD3 | 51133 | 8 | 1 | 34 | |
| GSE46452 | KCTD3 | 51133 | 3 | 1 | 55 | |
| GSE47630 | KCTD3 | 51133 | 15 | 0 | 25 | |
| GSE54993 | KCTD3 | 51133 | 0 | 6 | 64 | |
| GSE54994 | KCTD3 | 51133 | 14 | 0 | 39 | |
| GSE60625 | KCTD3 | 51133 | 0 | 0 | 11 | |
| GSE74703 | KCTD3 | 51133 | 8 | 1 | 27 | |
| GSE74704 | KCTD3 | 51133 | 3 | 0 | 17 | |
| TCGA | KCTD3 | 51133 | 43 | 3 | 50 |
Total number of gains: 112; Total number of losses: 12; Total Number of normals: 364.
Somatic mutations of KCTD3:
Generating mutation plots.
Highly correlated genes for KCTD3:
Showing top 20/1350 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCTD3 | SLC10A5 | 0.783975 | 3 | 0 | 3 |
| KCTD3 | MPP7 | 0.767136 | 7 | 0 | 7 |
| KCTD3 | GALNT12 | 0.756026 | 11 | 0 | 11 |
| KCTD3 | CAPN14 | 0.751819 | 6 | 0 | 6 |
| KCTD3 | GBP6 | 0.749813 | 4 | 0 | 4 |
| KCTD3 | WDR26 | 0.743304 | 10 | 0 | 10 |
| KCTD3 | LNX1 | 0.742026 | 7 | 0 | 6 |
| KCTD3 | CA13 | 0.740119 | 3 | 0 | 3 |
| KCTD3 | VSIG10L | 0.737963 | 6 | 0 | 6 |
| KCTD3 | SPINK7 | 0.736198 | 6 | 0 | 6 |
| KCTD3 | CASC3 | 0.735683 | 3 | 0 | 3 |
| KCTD3 | STX19 | 0.734255 | 6 | 0 | 6 |
| KCTD3 | HPGD | 0.730685 | 10 | 0 | 9 |
| KCTD3 | HDHD2 | 0.728759 | 4 | 0 | 4 |
| KCTD3 | UBL3 | 0.724456 | 11 | 0 | 11 |
| KCTD3 | FAM214A | 0.724211 | 6 | 0 | 5 |
| KCTD3 | CYSRT1 | 0.723614 | 6 | 0 | 6 |
| KCTD3 | GOLGA4 | 0.723051 | 8 | 0 | 7 |
| KCTD3 | KLB | 0.717875 | 5 | 0 | 5 |
| KCTD3 | MUC20 | 0.716296 | 6 | 0 | 6 |
For details and further investigation, click here