| Full name: kelch like family member 29 | Alias Symbol: KIAA1921 | ||
| Type: protein-coding gene | Cytoband: 2p24.1 | ||
| Entrez ID: 114818 | HGNC ID: HGNC:29404 | Ensembl Gene: ENSG00000119771 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of KLHL29:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KLHL29 | 114818 | 229310_at | -1.0836 | 0.0853 | |
| GSE20347 | KLHL29 | 114818 | 220348_at | -0.0264 | 0.8546 | |
| GSE23400 | KLHL29 | 114818 | 220348_at | -0.1066 | 0.0058 | |
| GSE26886 | KLHL29 | 114818 | 229310_at | 0.7170 | 0.1066 | |
| GSE29001 | KLHL29 | 114818 | 220348_at | -0.2127 | 0.3832 | |
| GSE38129 | KLHL29 | 114818 | 220348_at | -0.1535 | 0.2528 | |
| GSE45670 | KLHL29 | 114818 | 229310_at | -0.4580 | 0.1557 | |
| GSE53622 | KLHL29 | 114818 | 144378 | -0.0643 | 0.5836 | |
| GSE53624 | KLHL29 | 114818 | 23128 | -0.2605 | 0.0307 | |
| GSE63941 | KLHL29 | 114818 | 229310_at | -2.0391 | 0.1013 | |
| GSE77861 | KLHL29 | 114818 | 229310_at | 0.2340 | 0.6129 | |
| GSE97050 | KLHL29 | 114818 | A_33_P3359308 | -0.3612 | 0.2379 | |
| SRP064894 | KLHL29 | 114818 | RNAseq | -0.2818 | 0.4160 | |
| SRP133303 | KLHL29 | 114818 | RNAseq | -0.2404 | 0.3100 | |
| SRP159526 | KLHL29 | 114818 | RNAseq | 0.7763 | 0.0491 | |
| SRP193095 | KLHL29 | 114818 | RNAseq | 0.3904 | 0.0311 | |
| SRP219564 | KLHL29 | 114818 | RNAseq | 0.1949 | 0.7005 | |
| TCGA | KLHL29 | 114818 | RNAseq | 0.1213 | 0.3965 |
Upregulated datasets: 0; Downregulated datasets: 0.
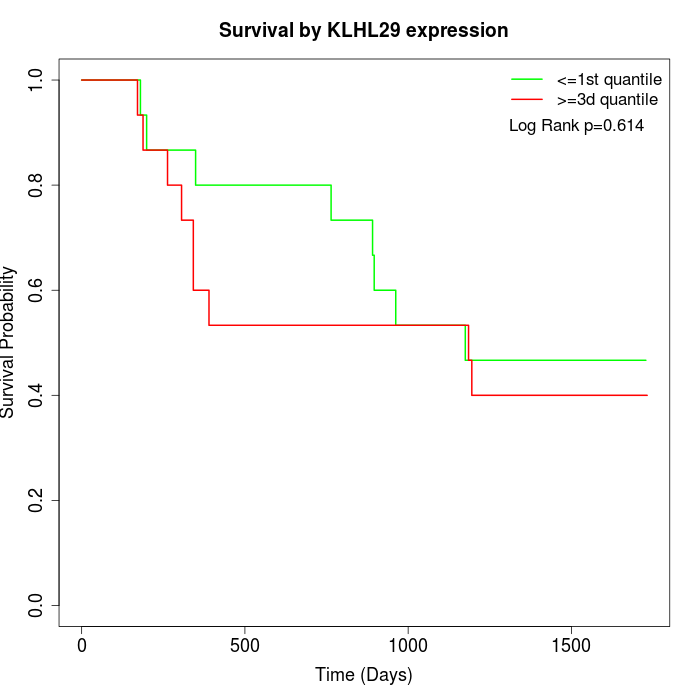
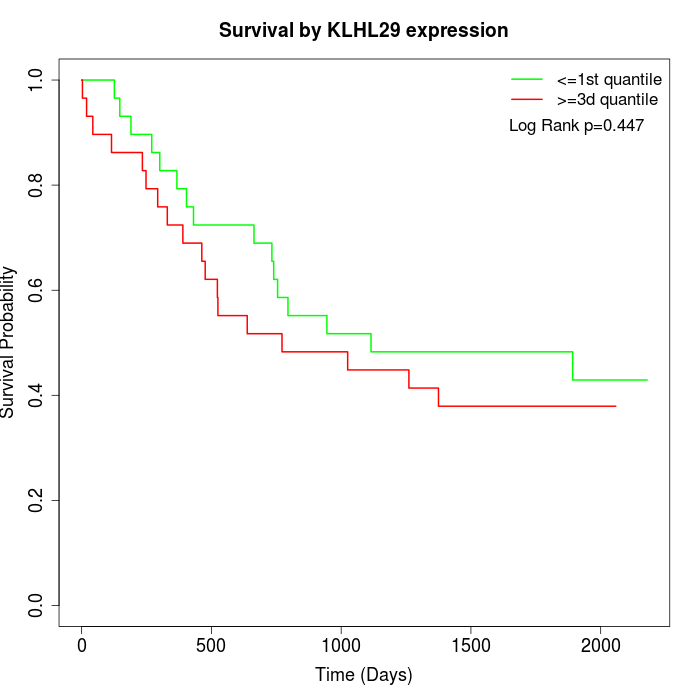
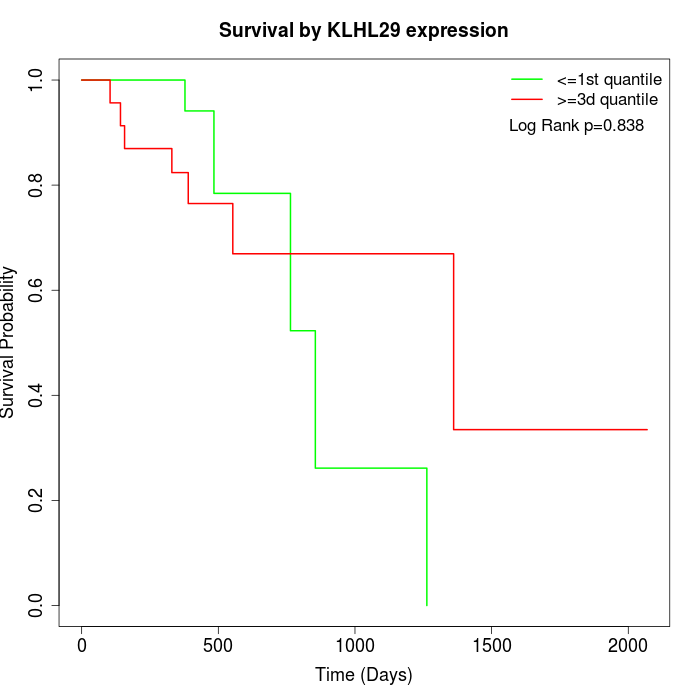
Survival by KLHL29 expression:
Note: Click image to view full size file.
Copy number change of KLHL29:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KLHL29 | 114818 | 10 | 2 | 18 | |
| GSE20123 | KLHL29 | 114818 | 10 | 2 | 18 | |
| GSE43470 | KLHL29 | 114818 | 2 | 1 | 40 | |
| GSE46452 | KLHL29 | 114818 | 3 | 4 | 52 | |
| GSE47630 | KLHL29 | 114818 | 8 | 0 | 32 | |
| GSE54993 | KLHL29 | 114818 | 0 | 7 | 63 | |
| GSE54994 | KLHL29 | 114818 | 10 | 0 | 43 | |
| GSE60625 | KLHL29 | 114818 | 0 | 3 | 8 | |
| GSE74703 | KLHL29 | 114818 | 2 | 0 | 34 | |
| GSE74704 | KLHL29 | 114818 | 9 | 1 | 10 | |
| TCGA | KLHL29 | 114818 | 35 | 3 | 58 |
Total number of gains: 89; Total number of losses: 23; Total Number of normals: 376.
Somatic mutations of KLHL29:
Generating mutation plots.
Highly correlated genes for KLHL29:
Showing top 20/254 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KLHL29 | SRR | 0.807335 | 3 | 0 | 3 |
| KLHL29 | MZT2B | 0.79232 | 3 | 0 | 3 |
| KLHL29 | POU6F1 | 0.773758 | 3 | 0 | 3 |
| KLHL29 | ZCCHC3 | 0.771642 | 3 | 0 | 3 |
| KLHL29 | ZNF721 | 0.766336 | 3 | 0 | 3 |
| KLHL29 | CTPS2 | 0.758725 | 3 | 0 | 3 |
| KLHL29 | ATP8B1 | 0.756981 | 3 | 0 | 3 |
| KLHL29 | ZXDA | 0.754598 | 3 | 0 | 3 |
| KLHL29 | RSF1 | 0.748101 | 3 | 0 | 3 |
| KLHL29 | DCLRE1C | 0.730975 | 3 | 0 | 3 |
| KLHL29 | COMMD6 | 0.726782 | 3 | 0 | 3 |
| KLHL29 | RPAP1 | 0.725454 | 3 | 0 | 3 |
| KLHL29 | GNE | 0.722065 | 3 | 0 | 3 |
| KLHL29 | SLC40A1 | 0.717567 | 3 | 0 | 3 |
| KLHL29 | RASSF3 | 0.713887 | 3 | 0 | 3 |
| KLHL29 | LATS2 | 0.710393 | 3 | 0 | 3 |
| KLHL29 | DZIP1 | 0.692765 | 3 | 0 | 3 |
| KLHL29 | MACROD2 | 0.689575 | 4 | 0 | 4 |
| KLHL29 | YPEL2 | 0.68829 | 3 | 0 | 3 |
| KLHL29 | VEZT | 0.686484 | 4 | 0 | 3 |
For details and further investigation, click here