| Full name: DNA cross-link repair 1C | Alias Symbol: ARTEMIS|FLJ11360|SNM1C|A-SCID | ||
| Type: protein-coding gene | Cytoband: 10p13 | ||
| Entrez ID: 64421 | HGNC ID: HGNC:17642 | Ensembl Gene: ENSG00000152457 | OMIM ID: 605988 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of DCLRE1C:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DCLRE1C | 64421 | 219678_x_at | -0.1038 | 0.8593 | |
| GSE20347 | DCLRE1C | 64421 | 219678_x_at | 0.2250 | 0.2370 | |
| GSE23400 | DCLRE1C | 64421 | 219678_x_at | -0.0364 | 0.5848 | |
| GSE26886 | DCLRE1C | 64421 | 219678_x_at | 0.3338 | 0.0953 | |
| GSE29001 | DCLRE1C | 64421 | 219678_x_at | 0.0188 | 0.9559 | |
| GSE38129 | DCLRE1C | 64421 | 219678_x_at | 0.0772 | 0.6801 | |
| GSE45670 | DCLRE1C | 64421 | 219678_x_at | 0.0699 | 0.6172 | |
| GSE53622 | DCLRE1C | 64421 | 63255 | -0.1920 | 0.0444 | |
| GSE53624 | DCLRE1C | 64421 | 63255 | -0.3297 | 0.0000 | |
| GSE63941 | DCLRE1C | 64421 | 219678_x_at | 0.0703 | 0.8635 | |
| GSE77861 | DCLRE1C | 64421 | 219678_x_at | 0.1621 | 0.6478 | |
| GSE97050 | DCLRE1C | 64421 | A_23_P86632 | 0.1440 | 0.7225 | |
| SRP007169 | DCLRE1C | 64421 | RNAseq | -0.2502 | 0.5119 | |
| SRP008496 | DCLRE1C | 64421 | RNAseq | -0.6438 | 0.0101 | |
| SRP064894 | DCLRE1C | 64421 | RNAseq | -0.2925 | 0.0596 | |
| SRP133303 | DCLRE1C | 64421 | RNAseq | -0.2547 | 0.0529 | |
| SRP159526 | DCLRE1C | 64421 | RNAseq | -1.0598 | 0.0000 | |
| SRP193095 | DCLRE1C | 64421 | RNAseq | -0.2818 | 0.0008 | |
| SRP219564 | DCLRE1C | 64421 | RNAseq | -0.8316 | 0.0427 | |
| TCGA | DCLRE1C | 64421 | RNAseq | 0.0952 | 0.1386 |
Upregulated datasets: 0; Downregulated datasets: 1.
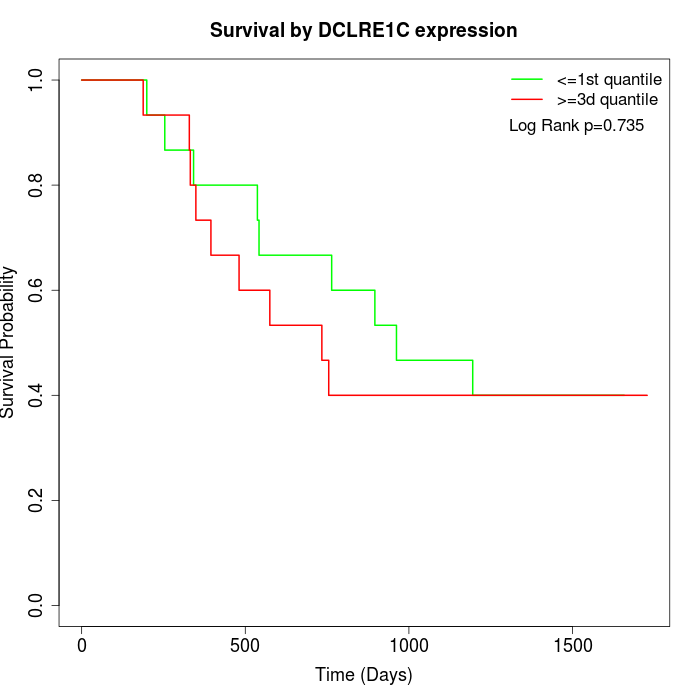
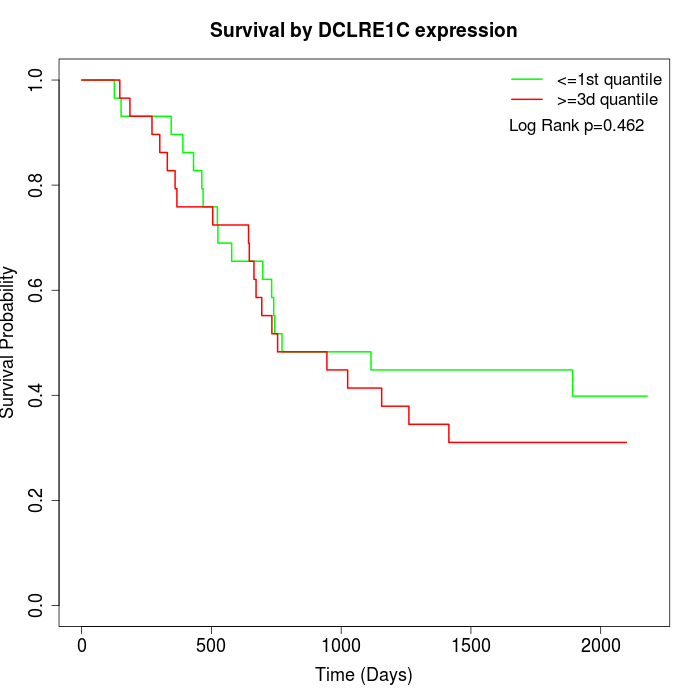
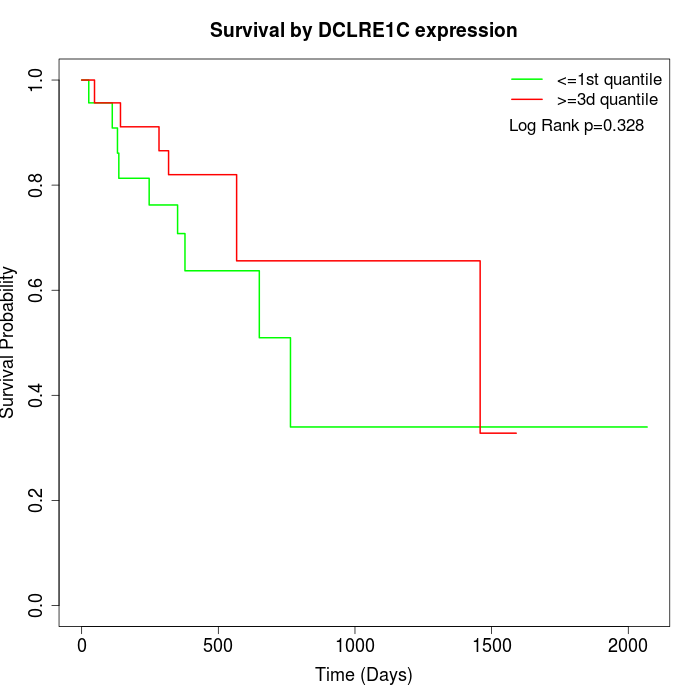
Survival by DCLRE1C expression:
Note: Click image to view full size file.
Copy number change of DCLRE1C:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DCLRE1C | 64421 | 3 | 8 | 19 | |
| GSE20123 | DCLRE1C | 64421 | 3 | 7 | 20 | |
| GSE43470 | DCLRE1C | 64421 | 3 | 5 | 35 | |
| GSE46452 | DCLRE1C | 64421 | 1 | 14 | 44 | |
| GSE47630 | DCLRE1C | 64421 | 5 | 15 | 20 | |
| GSE54993 | DCLRE1C | 64421 | 10 | 0 | 60 | |
| GSE54994 | DCLRE1C | 64421 | 4 | 9 | 40 | |
| GSE60625 | DCLRE1C | 64421 | 0 | 0 | 11 | |
| GSE74703 | DCLRE1C | 64421 | 2 | 3 | 31 | |
| GSE74704 | DCLRE1C | 64421 | 0 | 6 | 14 | |
| TCGA | DCLRE1C | 64421 | 20 | 25 | 51 |
Total number of gains: 51; Total number of losses: 92; Total Number of normals: 345.
Somatic mutations of DCLRE1C:
Generating mutation plots.
Highly correlated genes for DCLRE1C:
Showing top 20/224 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DCLRE1C | BIRC3 | 0.848946 | 3 | 0 | 3 |
| DCLRE1C | HAUS2 | 0.797171 | 9 | 0 | 9 |
| DCLRE1C | MAGI1 | 0.790483 | 4 | 0 | 4 |
| DCLRE1C | ALMS1 | 0.775238 | 8 | 0 | 8 |
| DCLRE1C | SLC22A3 | 0.773199 | 4 | 0 | 4 |
| DCLRE1C | SLC35E1 | 0.763233 | 10 | 0 | 9 |
| DCLRE1C | ERGIC1 | 0.757948 | 3 | 0 | 3 |
| DCLRE1C | PILRA | 0.753485 | 3 | 0 | 3 |
| DCLRE1C | ZNF611 | 0.750447 | 8 | 0 | 8 |
| DCLRE1C | LILRB1 | 0.749369 | 3 | 0 | 3 |
| DCLRE1C | C9orf64 | 0.747666 | 6 | 0 | 5 |
| DCLRE1C | RPL22L1 | 0.74717 | 3 | 0 | 3 |
| DCLRE1C | GTF2H3 | 0.745308 | 9 | 0 | 9 |
| DCLRE1C | ZNF721 | 0.741579 | 9 | 0 | 8 |
| DCLRE1C | CMBL | 0.739673 | 4 | 0 | 4 |
| DCLRE1C | FAM161B | 0.734721 | 5 | 0 | 5 |
| DCLRE1C | POLR1B | 0.734318 | 8 | 0 | 6 |
| DCLRE1C | MCM8 | 0.732798 | 3 | 0 | 3 |
| DCLRE1C | CCDC84 | 0.732167 | 4 | 0 | 4 |
| DCLRE1C | KLHL29 | 0.730975 | 3 | 0 | 3 |
For details and further investigation, click here