| Full name: large tumor suppressor kinase 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 13q12.11 | ||
| Entrez ID: 26524 | HGNC ID: HGNC:6515 | Ensembl Gene: ENSG00000150457 | OMIM ID: 604861 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
LATS2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04390 | Hippo signaling pathway |
Expression of LATS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LATS2 | 26524 | 227013_at | -0.6190 | 0.2775 | |
| GSE26886 | LATS2 | 26524 | 227013_at | -0.6495 | 0.0154 | |
| GSE45670 | LATS2 | 26524 | 227013_at | -0.8016 | 0.0000 | |
| GSE53622 | LATS2 | 26524 | 44500 | -0.4197 | 0.0000 | |
| GSE53624 | LATS2 | 26524 | 44500 | -0.5835 | 0.0000 | |
| GSE63941 | LATS2 | 26524 | 223380_s_at | -1.5215 | 0.0092 | |
| GSE77861 | LATS2 | 26524 | 227013_at | -0.5512 | 0.1566 | |
| GSE97050 | LATS2 | 26524 | A_24_P70002 | -0.6118 | 0.1387 | |
| SRP007169 | LATS2 | 26524 | RNAseq | -1.4243 | 0.0011 | |
| SRP008496 | LATS2 | 26524 | RNAseq | -0.8079 | 0.0068 | |
| SRP064894 | LATS2 | 26524 | RNAseq | -0.5680 | 0.0520 | |
| SRP133303 | LATS2 | 26524 | RNAseq | -0.3575 | 0.1073 | |
| SRP159526 | LATS2 | 26524 | RNAseq | -0.7211 | 0.0154 | |
| SRP193095 | LATS2 | 26524 | RNAseq | -0.5310 | 0.0106 | |
| SRP219564 | LATS2 | 26524 | RNAseq | -0.4958 | 0.2683 | |
| TCGA | LATS2 | 26524 | RNAseq | -0.2455 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 2.
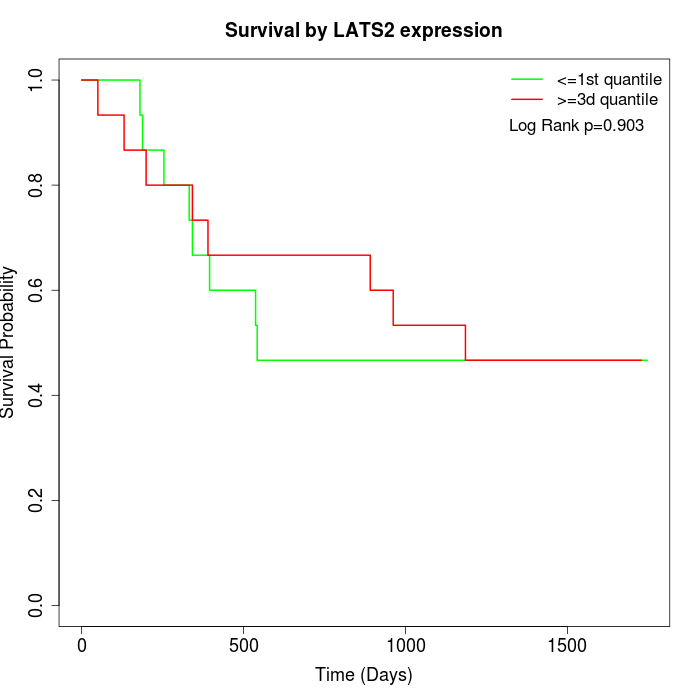
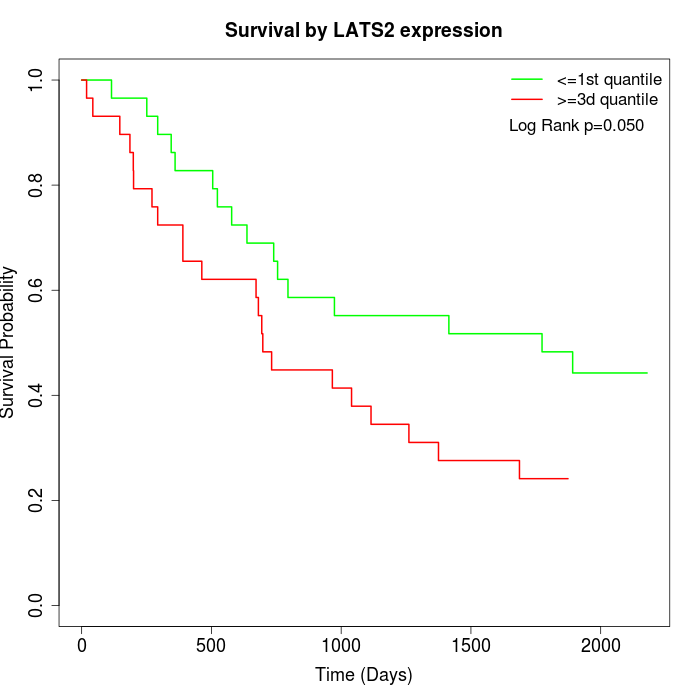
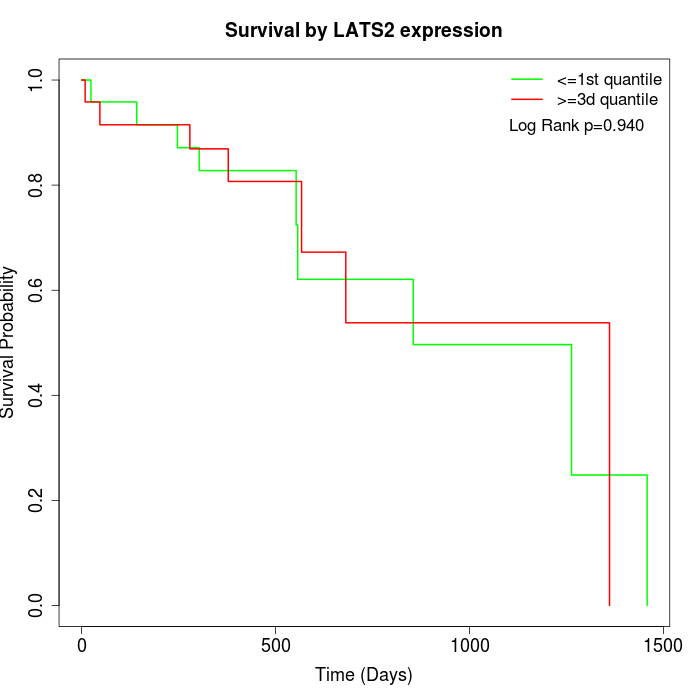
Survival by LATS2 expression:
Note: Click image to view full size file.
Copy number change of LATS2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LATS2 | 26524 | 1 | 15 | 14 | |
| GSE20123 | LATS2 | 26524 | 1 | 14 | 15 | |
| GSE43470 | LATS2 | 26524 | 2 | 13 | 28 | |
| GSE46452 | LATS2 | 26524 | 0 | 33 | 26 | |
| GSE47630 | LATS2 | 26524 | 3 | 26 | 11 | |
| GSE54993 | LATS2 | 26524 | 11 | 2 | 57 | |
| GSE54994 | LATS2 | 26524 | 1 | 13 | 39 | |
| GSE60625 | LATS2 | 26524 | 0 | 3 | 8 | |
| GSE74703 | LATS2 | 26524 | 2 | 10 | 24 | |
| GSE74704 | LATS2 | 26524 | 0 | 12 | 8 | |
| TCGA | LATS2 | 26524 | 6 | 43 | 47 |
Total number of gains: 27; Total number of losses: 184; Total Number of normals: 277.
Somatic mutations of LATS2:
Generating mutation plots.
Highly correlated genes for LATS2:
Showing top 20/897 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LATS2 | DPH3 | 0.767942 | 3 | 0 | 3 |
| LATS2 | MYO9A | 0.747115 | 4 | 0 | 4 |
| LATS2 | ALDH1A2 | 0.729152 | 4 | 0 | 4 |
| LATS2 | CLCC1 | 0.72659 | 4 | 0 | 3 |
| LATS2 | ZC3H13 | 0.725545 | 3 | 0 | 3 |
| LATS2 | ILK | 0.721874 | 6 | 0 | 5 |
| LATS2 | USP9X | 0.716756 | 3 | 0 | 3 |
| LATS2 | KLHL29 | 0.710393 | 3 | 0 | 3 |
| LATS2 | CACNA1C | 0.70897 | 5 | 0 | 5 |
| LATS2 | OR7A10 | 0.708731 | 3 | 0 | 3 |
| LATS2 | SST | 0.705588 | 3 | 0 | 3 |
| LATS2 | SOX17 | 0.704277 | 4 | 0 | 4 |
| LATS2 | ARHGAP31 | 0.70362 | 5 | 0 | 5 |
| LATS2 | PRICKLE2 | 0.70257 | 7 | 0 | 7 |
| LATS2 | COX7A1 | 0.70218 | 6 | 0 | 6 |
| LATS2 | LRCH1 | 0.69997 | 4 | 0 | 4 |
| LATS2 | USP12 | 0.696984 | 4 | 0 | 3 |
| LATS2 | SGCA | 0.695079 | 5 | 0 | 5 |
| LATS2 | MAGI1 | 0.693193 | 4 | 0 | 4 |
| LATS2 | FIBIN | 0.692894 | 5 | 0 | 4 |
For details and further investigation, click here