| Full name: RNA polymerase II associated protein 1 | Alias Symbol: DKFZP727M111|KIAA1403|MGC858|FLJ12732 | ||
| Type: protein-coding gene | Cytoband: 15q15.1 | ||
| Entrez ID: 26015 | HGNC ID: HGNC:24567 | Ensembl Gene: ENSG00000103932 | OMIM ID: 611475 |
| Drug and gene relationship at DGIdb | |||
Expression of RPAP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RPAP1 | 26015 | 218441_s_at | 0.2141 | 0.6068 | |
| GSE20347 | RPAP1 | 26015 | 218441_s_at | 0.1810 | 0.0324 | |
| GSE23400 | RPAP1 | 26015 | 218441_s_at | 0.1434 | 0.0008 | |
| GSE26886 | RPAP1 | 26015 | 218441_s_at | 0.2336 | 0.1020 | |
| GSE29001 | RPAP1 | 26015 | 218441_s_at | 0.0597 | 0.7866 | |
| GSE38129 | RPAP1 | 26015 | 218441_s_at | 0.0820 | 0.4362 | |
| GSE45670 | RPAP1 | 26015 | 218441_s_at | 0.2970 | 0.0420 | |
| GSE53622 | RPAP1 | 26015 | 98785 | 0.0966 | 0.1734 | |
| GSE53624 | RPAP1 | 26015 | 98785 | 0.1679 | 0.0388 | |
| GSE63941 | RPAP1 | 26015 | 218441_s_at | 0.2822 | 0.4272 | |
| GSE77861 | RPAP1 | 26015 | 218441_s_at | 0.2234 | 0.3884 | |
| GSE97050 | RPAP1 | 26015 | A_23_P3453 | -0.2082 | 0.2705 | |
| SRP007169 | RPAP1 | 26015 | RNAseq | 0.0388 | 0.9155 | |
| SRP008496 | RPAP1 | 26015 | RNAseq | -0.2907 | 0.3962 | |
| SRP064894 | RPAP1 | 26015 | RNAseq | 0.2289 | 0.0773 | |
| SRP133303 | RPAP1 | 26015 | RNAseq | 0.0740 | 0.6195 | |
| SRP159526 | RPAP1 | 26015 | RNAseq | 0.2492 | 0.3597 | |
| SRP193095 | RPAP1 | 26015 | RNAseq | 0.3126 | 0.0013 | |
| SRP219564 | RPAP1 | 26015 | RNAseq | -0.0918 | 0.6594 | |
| TCGA | RPAP1 | 26015 | RNAseq | 0.0114 | 0.8212 |
Upregulated datasets: 0; Downregulated datasets: 0.
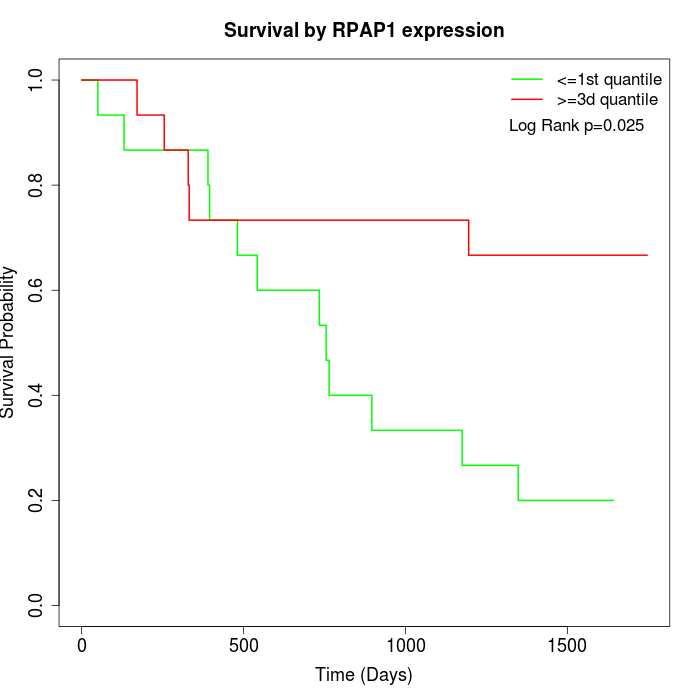
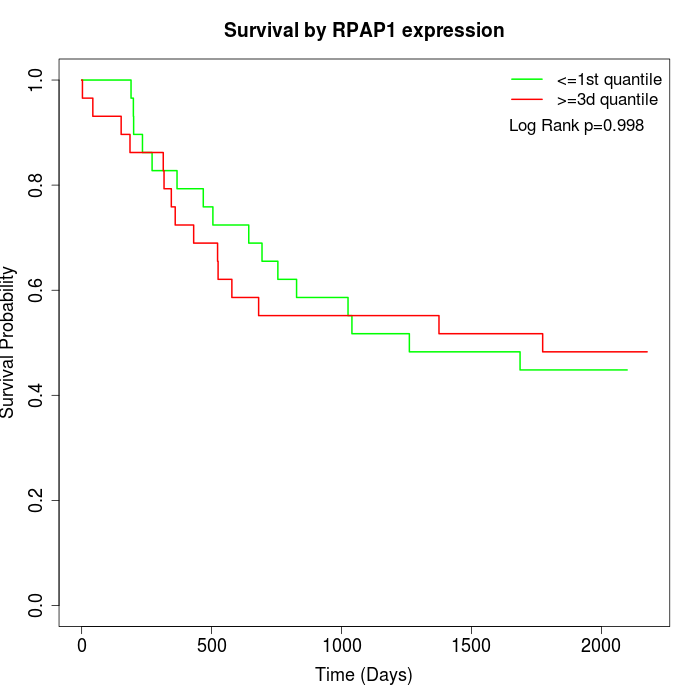
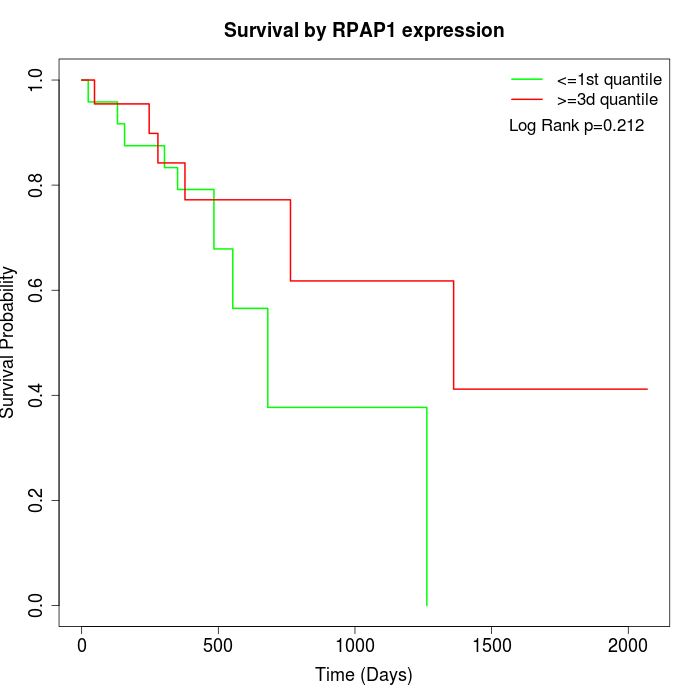
Survival by RPAP1 expression:
Note: Click image to view full size file.
Copy number change of RPAP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RPAP1 | 26015 | 4 | 5 | 21 | |
| GSE20123 | RPAP1 | 26015 | 4 | 5 | 21 | |
| GSE43470 | RPAP1 | 26015 | 4 | 5 | 34 | |
| GSE46452 | RPAP1 | 26015 | 3 | 7 | 49 | |
| GSE47630 | RPAP1 | 26015 | 8 | 11 | 21 | |
| GSE54993 | RPAP1 | 26015 | 4 | 6 | 60 | |
| GSE54994 | RPAP1 | 26015 | 5 | 7 | 41 | |
| GSE60625 | RPAP1 | 26015 | 4 | 0 | 7 | |
| GSE74703 | RPAP1 | 26015 | 4 | 4 | 28 | |
| GSE74704 | RPAP1 | 26015 | 3 | 3 | 14 | |
| TCGA | RPAP1 | 26015 | 11 | 18 | 67 |
Total number of gains: 54; Total number of losses: 71; Total Number of normals: 363.
Somatic mutations of RPAP1:
Generating mutation plots.
Highly correlated genes for RPAP1:
Showing top 20/274 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RPAP1 | FAM89B | 0.80968 | 3 | 0 | 3 |
| RPAP1 | GATC | 0.729852 | 3 | 0 | 3 |
| RPAP1 | KLHL29 | 0.725454 | 3 | 0 | 3 |
| RPAP1 | RBBP4 | 0.715966 | 3 | 0 | 3 |
| RPAP1 | C11orf1 | 0.715471 | 3 | 0 | 3 |
| RPAP1 | BAZ1B | 0.707569 | 4 | 0 | 4 |
| RPAP1 | IRS2 | 0.695201 | 3 | 0 | 3 |
| RPAP1 | QSER1 | 0.694683 | 3 | 0 | 3 |
| RPAP1 | ORAI1 | 0.692191 | 4 | 0 | 4 |
| RPAP1 | FOXRED2 | 0.691374 | 3 | 0 | 3 |
| RPAP1 | FAR2 | 0.691304 | 3 | 0 | 3 |
| RPAP1 | PLCB4 | 0.690495 | 3 | 0 | 3 |
| RPAP1 | ZXDB | 0.688285 | 3 | 0 | 3 |
| RPAP1 | MOGS | 0.682194 | 5 | 0 | 5 |
| RPAP1 | NADSYN1 | 0.682068 | 3 | 0 | 3 |
| RPAP1 | NLK | 0.678347 | 3 | 0 | 3 |
| RPAP1 | MSH2 | 0.677574 | 4 | 0 | 4 |
| RPAP1 | KCTD1 | 0.673663 | 3 | 0 | 3 |
| RPAP1 | NME4 | 0.671619 | 4 | 0 | 3 |
| RPAP1 | CRTC3 | 0.668828 | 3 | 0 | 3 |
For details and further investigation, click here