| Full name: keratin associated protein 4-1 | Alias Symbol: KAP4.1|KAP4.10 | ||
| Type: protein-coding gene | Cytoband: 17q21.2 | ||
| Entrez ID: 85285 | HGNC ID: HGNC:18907 | Ensembl Gene: ENSG00000198443 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of KRTAP4-1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KRTAP4-1 | 85285 | 234635_at | 0.6149 | 0.0498 | |
| GSE26886 | KRTAP4-1 | 85285 | 234635_at | 1.1743 | 0.0012 | |
| GSE45670 | KRTAP4-1 | 85285 | 234635_at | 1.0350 | 0.0046 | |
| GSE53622 | KRTAP4-1 | 85285 | 64937 | -0.8566 | 0.0000 | |
| GSE53624 | KRTAP4-1 | 85285 | 64937 | -2.5039 | 0.0000 | |
| GSE63941 | KRTAP4-1 | 85285 | 234635_at | 1.5827 | 0.1585 | |
| GSE77861 | KRTAP4-1 | 85285 | 234635_at | 0.4961 | 0.0558 | |
| GSE97050 | KRTAP4-1 | 85285 | A_33_P3245348 | -0.5359 | 0.2513 | |
| TCGA | KRTAP4-1 | 85285 | RNAseq | 6.1238 | 0.0000 |
Upregulated datasets: 3; Downregulated datasets: 1.
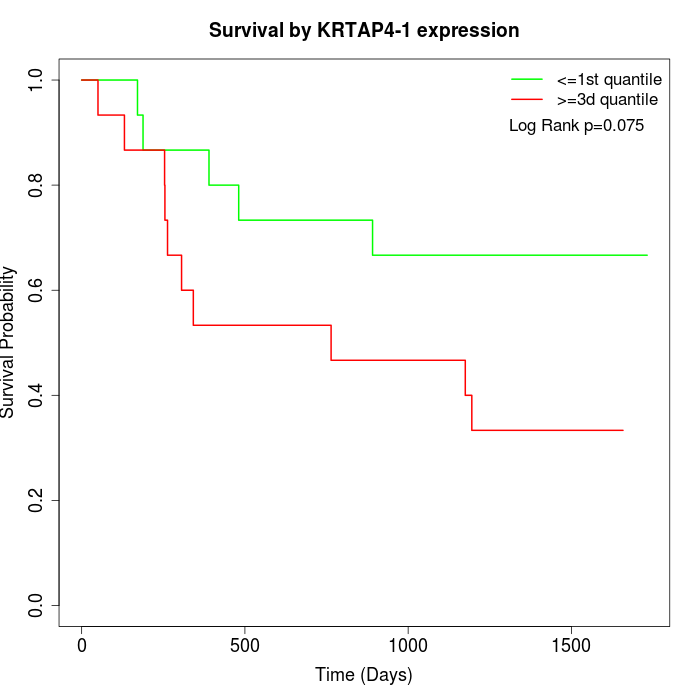
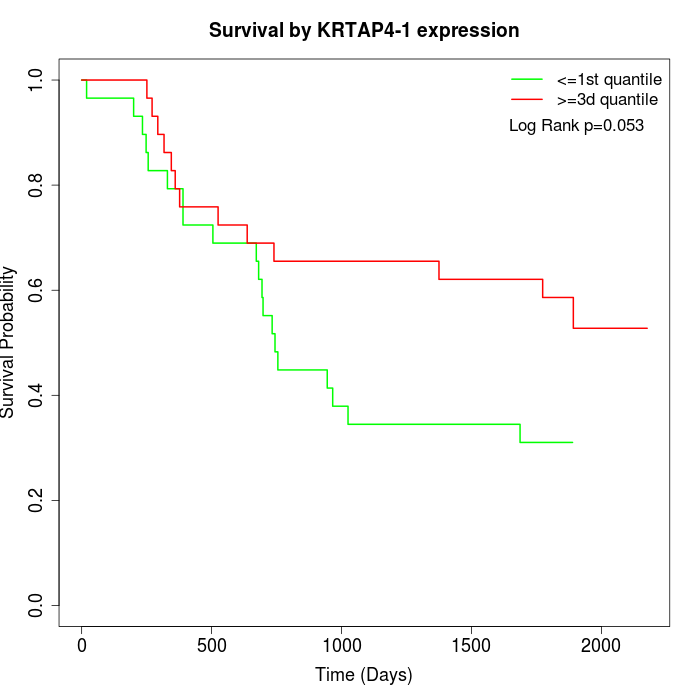
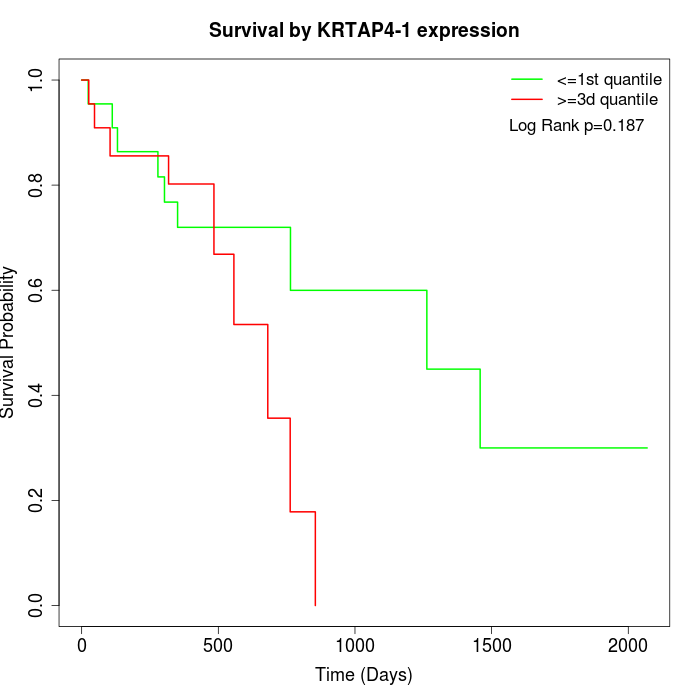
Survival by KRTAP4-1 expression:
Note: Click image to view full size file.
Copy number change of KRTAP4-1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KRTAP4-1 | 85285 | 7 | 1 | 22 | |
| GSE20123 | KRTAP4-1 | 85285 | 7 | 1 | 22 | |
| GSE43470 | KRTAP4-1 | 85285 | 1 | 2 | 40 | |
| GSE46452 | KRTAP4-1 | 85285 | 34 | 0 | 25 | |
| GSE47630 | KRTAP4-1 | 85285 | 8 | 1 | 31 | |
| GSE54993 | KRTAP4-1 | 85285 | 3 | 4 | 63 | |
| GSE54994 | KRTAP4-1 | 85285 | 8 | 5 | 40 | |
| GSE60625 | KRTAP4-1 | 85285 | 4 | 0 | 7 | |
| GSE74703 | KRTAP4-1 | 85285 | 1 | 1 | 34 | |
| GSE74704 | KRTAP4-1 | 85285 | 4 | 1 | 15 |
Total number of gains: 77; Total number of losses: 16; Total Number of normals: 299.
Somatic mutations of KRTAP4-1:
Generating mutation plots.
Highly correlated genes for KRTAP4-1:
Showing top 20/206 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KRTAP4-1 | KRTAP20-1 | 0.853859 | 3 | 0 | 3 |
| KRTAP4-1 | BARHL2 | 0.838144 | 3 | 0 | 3 |
| KRTAP4-1 | FAM163B | 0.835446 | 3 | 0 | 3 |
| KRTAP4-1 | PTCH2 | 0.831313 | 3 | 0 | 3 |
| KRTAP4-1 | MFSD2B | 0.831205 | 3 | 0 | 3 |
| KRTAP4-1 | OR10G8 | 0.826254 | 3 | 0 | 3 |
| KRTAP4-1 | TEN1 | 0.790766 | 3 | 0 | 3 |
| KRTAP4-1 | SLC52A1 | 0.78754 | 3 | 0 | 3 |
| KRTAP4-1 | KRTAP6-1 | 0.785344 | 3 | 0 | 3 |
| KRTAP4-1 | RRP1 | 0.780909 | 4 | 0 | 4 |
| KRTAP4-1 | SEC14L4 | 0.776175 | 3 | 0 | 3 |
| KRTAP4-1 | CD164L2 | 0.77592 | 3 | 0 | 3 |
| KRTAP4-1 | OR2Z1 | 0.771109 | 4 | 0 | 3 |
| KRTAP4-1 | TOX2 | 0.769595 | 3 | 0 | 3 |
| KRTAP4-1 | PKD2L1 | 0.7678 | 3 | 0 | 3 |
| KRTAP4-1 | RNF151 | 0.766767 | 3 | 0 | 3 |
| KRTAP4-1 | TECPR1 | 0.75578 | 3 | 0 | 3 |
| KRTAP4-1 | PRR18 | 0.75459 | 3 | 0 | 3 |
| KRTAP4-1 | KRTAP5-4 | 0.753388 | 3 | 0 | 3 |
| KRTAP4-1 | OR10G3 | 0.750128 | 3 | 0 | 3 |
For details and further investigation, click here