| Full name: late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 | Alias Symbol: MAPBPIP|MAPKSP1AP|p14|ENDAP|Ragulator2 | ||
| Type: protein-coding gene | Cytoband: 1q22 | ||
| Entrez ID: 28956 | HGNC ID: HGNC:29796 | Ensembl Gene: ENSG00000116586 | OMIM ID: 610389 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
LAMTOR2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04150 | mTOR signaling pathway |
Expression of LAMTOR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LAMTOR2 | 28956 | 218291_at | 0.1683 | 0.5675 | |
| GSE20347 | LAMTOR2 | 28956 | 218291_at | -0.4655 | 0.0011 | |
| GSE23400 | LAMTOR2 | 28956 | 218291_at | -0.0337 | 0.5860 | |
| GSE26886 | LAMTOR2 | 28956 | 218291_at | 0.4737 | 0.0137 | |
| GSE29001 | LAMTOR2 | 28956 | 218291_at | -0.0794 | 0.7700 | |
| GSE38129 | LAMTOR2 | 28956 | 218291_at | -0.1547 | 0.3174 | |
| GSE45670 | LAMTOR2 | 28956 | 218291_at | 0.0055 | 0.9678 | |
| GSE53622 | LAMTOR2 | 28956 | 54734 | -0.1002 | 0.0560 | |
| GSE53624 | LAMTOR2 | 28956 | 54734 | 0.0071 | 0.9118 | |
| GSE63941 | LAMTOR2 | 28956 | 218291_at | -0.2554 | 0.4158 | |
| GSE77861 | LAMTOR2 | 28956 | 218291_at | -0.0764 | 0.7649 | |
| GSE97050 | LAMTOR2 | 28956 | A_33_P3246068 | 0.0217 | 0.9323 | |
| SRP007169 | LAMTOR2 | 28956 | RNAseq | -1.2003 | 0.0003 | |
| SRP008496 | LAMTOR2 | 28956 | RNAseq | -0.6867 | 0.0024 | |
| SRP064894 | LAMTOR2 | 28956 | RNAseq | 0.7455 | 0.0548 | |
| SRP133303 | LAMTOR2 | 28956 | RNAseq | 0.1922 | 0.1935 | |
| SRP159526 | LAMTOR2 | 28956 | RNAseq | -0.0164 | 0.9221 | |
| SRP193095 | LAMTOR2 | 28956 | RNAseq | -0.2029 | 0.0963 | |
| SRP219564 | LAMTOR2 | 28956 | RNAseq | -0.0482 | 0.8860 |
Upregulated datasets: 0; Downregulated datasets: 1.
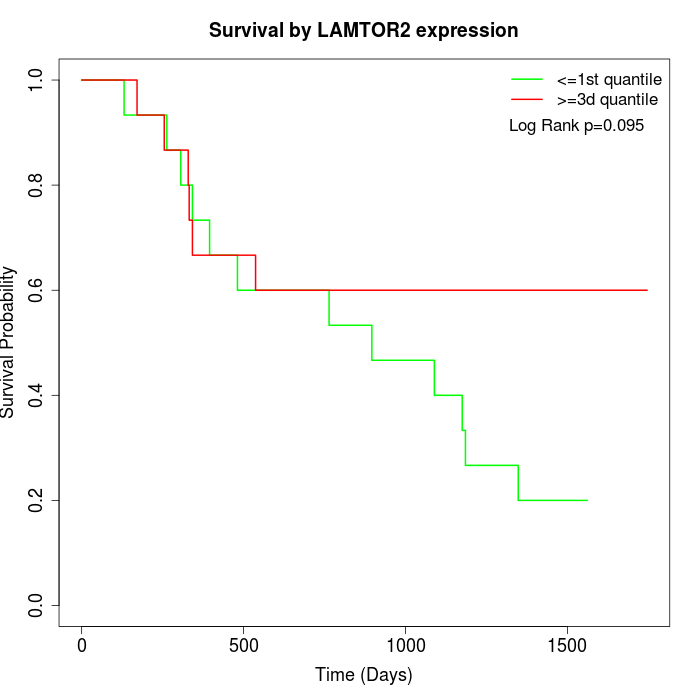
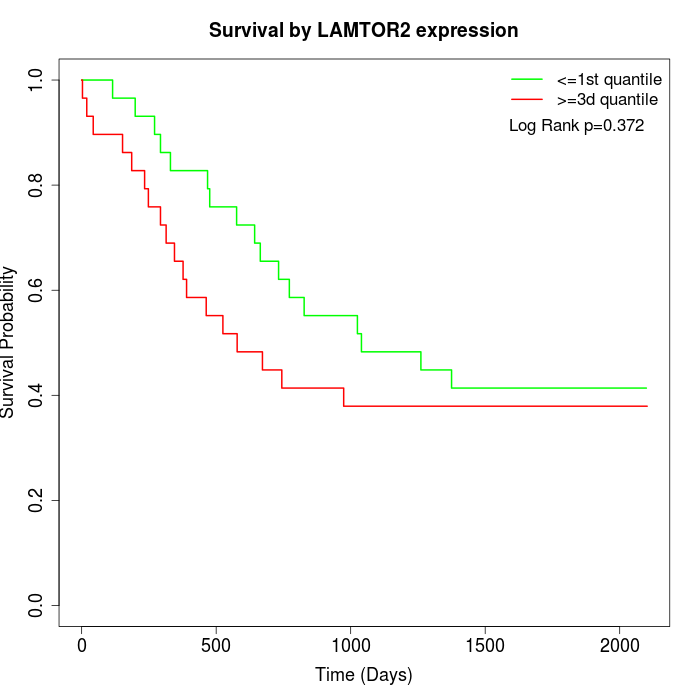
Survival by LAMTOR2 expression:
Note: Click image to view full size file.
Copy number change of LAMTOR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LAMTOR2 | 28956 | 14 | 0 | 16 | |
| GSE20123 | LAMTOR2 | 28956 | 13 | 0 | 17 | |
| GSE43470 | LAMTOR2 | 28956 | 7 | 2 | 34 | |
| GSE46452 | LAMTOR2 | 28956 | 2 | 1 | 56 | |
| GSE47630 | LAMTOR2 | 28956 | 14 | 0 | 26 | |
| GSE54993 | LAMTOR2 | 28956 | 0 | 5 | 65 | |
| GSE54994 | LAMTOR2 | 28956 | 16 | 0 | 37 | |
| GSE60625 | LAMTOR2 | 28956 | 0 | 0 | 11 | |
| GSE74703 | LAMTOR2 | 28956 | 7 | 2 | 27 | |
| GSE74704 | LAMTOR2 | 28956 | 6 | 0 | 14 | |
| TCGA | LAMTOR2 | 28956 | 38 | 2 | 56 |
Total number of gains: 117; Total number of losses: 12; Total Number of normals: 359.
Somatic mutations of LAMTOR2:
Generating mutation plots.
Highly correlated genes for LAMTOR2:
Showing top 20/100 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LAMTOR2 | AIFM3 | 0.752097 | 3 | 0 | 3 |
| LAMTOR2 | ZC3H10 | 0.74989 | 3 | 0 | 3 |
| LAMTOR2 | KLHL35 | 0.747694 | 3 | 0 | 3 |
| LAMTOR2 | C19orf81 | 0.738207 | 3 | 0 | 3 |
| LAMTOR2 | STK32C | 0.735989 | 3 | 0 | 3 |
| LAMTOR2 | JSRP1 | 0.733658 | 3 | 0 | 3 |
| LAMTOR2 | RAG2 | 0.719959 | 3 | 0 | 3 |
| LAMTOR2 | WDR60 | 0.715114 | 3 | 0 | 3 |
| LAMTOR2 | CLN3 | 0.708203 | 3 | 0 | 3 |
| LAMTOR2 | FUT10 | 0.707562 | 3 | 0 | 3 |
| LAMTOR2 | IL17RE | 0.705174 | 3 | 0 | 3 |
| LAMTOR2 | ETV3 | 0.70442 | 3 | 0 | 3 |
| LAMTOR2 | ZNF777 | 0.698397 | 5 | 0 | 4 |
| LAMTOR2 | WRAP73 | 0.695208 | 3 | 0 | 3 |
| LAMTOR2 | REG3A | 0.694623 | 3 | 0 | 3 |
| LAMTOR2 | STAC3 | 0.687653 | 3 | 0 | 3 |
| LAMTOR2 | IRAK1 | 0.687507 | 4 | 0 | 3 |
| LAMTOR2 | SDCCAG8 | 0.682146 | 3 | 0 | 3 |
| LAMTOR2 | ADPRHL2 | 0.679555 | 3 | 0 | 3 |
| LAMTOR2 | CCDC142 | 0.679131 | 3 | 0 | 3 |
For details and further investigation, click here