| Full name: lymphoid enhancer binding factor 1 | Alias Symbol: TCF1ALPHA|TCF10|TCF7L3 | ||
| Type: protein-coding gene | Cytoband: 4q25 | ||
| Entrez ID: 51176 | HGNC ID: HGNC:6551 | Ensembl Gene: ENSG00000138795 | OMIM ID: 153245 |
| Drug and gene relationship at DGIdb | |||
LEF1 involved pathways:
Expression of LEF1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LEF1 | 51176 | 221558_s_at | 0.2373 | 0.7018 | |
| GSE20347 | LEF1 | 51176 | 221558_s_at | 0.6272 | 0.0414 | |
| GSE23400 | LEF1 | 51176 | 221558_s_at | 0.6535 | 0.0000 | |
| GSE26886 | LEF1 | 51176 | 221558_s_at | 0.8039 | 0.1188 | |
| GSE29001 | LEF1 | 51176 | 221558_s_at | 0.0144 | 0.9829 | |
| GSE38129 | LEF1 | 51176 | 221558_s_at | 1.0181 | 0.0004 | |
| GSE45670 | LEF1 | 51176 | 221558_s_at | 0.3822 | 0.1438 | |
| GSE53622 | LEF1 | 51176 | 23039 | 0.9370 | 0.0000 | |
| GSE53624 | LEF1 | 51176 | 21090 | 0.5997 | 0.0000 | |
| GSE63941 | LEF1 | 51176 | 221558_s_at | -0.6444 | 0.6602 | |
| GSE77861 | LEF1 | 51176 | 221558_s_at | 0.9111 | 0.0392 | |
| GSE97050 | LEF1 | 51176 | A_24_P20630 | 0.7827 | 0.1090 | |
| SRP007169 | LEF1 | 51176 | RNAseq | 0.4939 | 0.3773 | |
| SRP008496 | LEF1 | 51176 | RNAseq | 1.0991 | 0.0063 | |
| SRP064894 | LEF1 | 51176 | RNAseq | 0.7680 | 0.0014 | |
| SRP133303 | LEF1 | 51176 | RNAseq | 0.6537 | 0.0344 | |
| SRP159526 | LEF1 | 51176 | RNAseq | 1.3992 | 0.0018 | |
| SRP193095 | LEF1 | 51176 | RNAseq | 0.3870 | 0.1276 | |
| SRP219564 | LEF1 | 51176 | RNAseq | 0.7542 | 0.2254 | |
| TCGA | LEF1 | 51176 | RNAseq | 0.6776 | 0.0000 |
Upregulated datasets: 3; Downregulated datasets: 0.
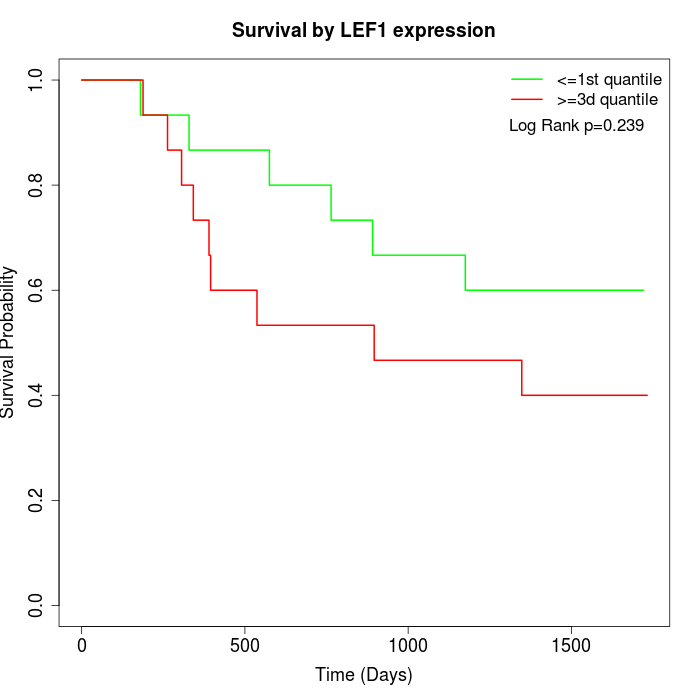
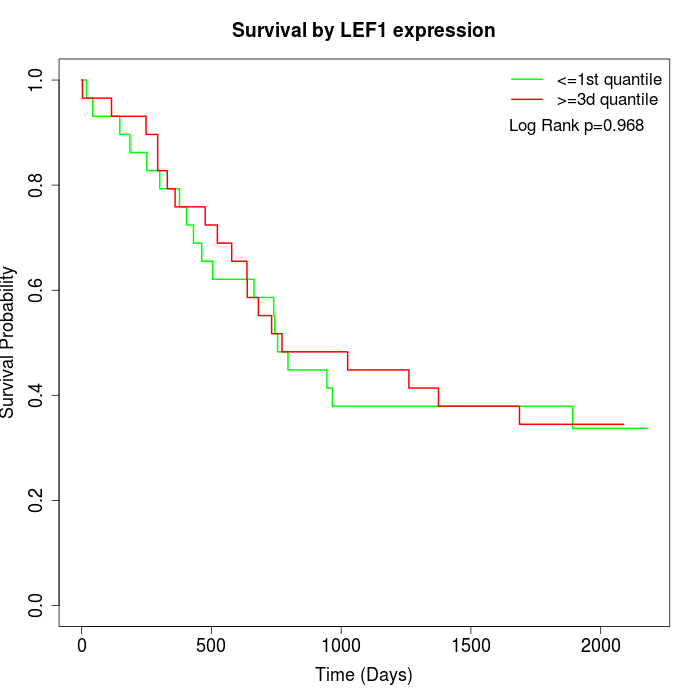
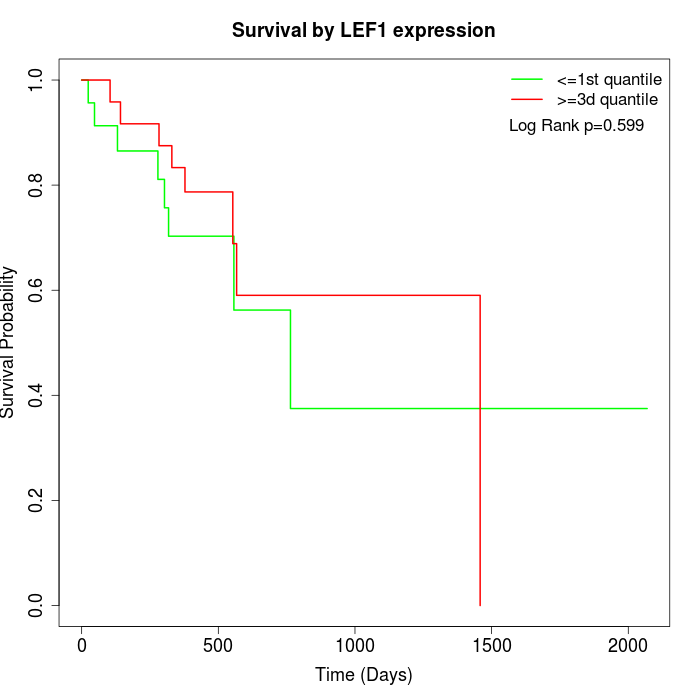
Survival by LEF1 expression:
Note: Click image to view full size file.
Copy number change of LEF1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LEF1 | 51176 | 0 | 12 | 18 | |
| GSE20123 | LEF1 | 51176 | 0 | 12 | 18 | |
| GSE43470 | LEF1 | 51176 | 0 | 13 | 30 | |
| GSE46452 | LEF1 | 51176 | 1 | 36 | 22 | |
| GSE47630 | LEF1 | 51176 | 0 | 20 | 20 | |
| GSE54993 | LEF1 | 51176 | 9 | 0 | 61 | |
| GSE54994 | LEF1 | 51176 | 1 | 10 | 42 | |
| GSE60625 | LEF1 | 51176 | 0 | 3 | 8 | |
| GSE74703 | LEF1 | 51176 | 0 | 11 | 25 | |
| GSE74704 | LEF1 | 51176 | 0 | 6 | 14 | |
| TCGA | LEF1 | 51176 | 8 | 35 | 53 |
Total number of gains: 19; Total number of losses: 158; Total Number of normals: 311.
Somatic mutations of LEF1:
Generating mutation plots.
Highly correlated genes for LEF1:
Showing top 20/570 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LEF1 | POU5F2 | 0.765156 | 3 | 0 | 3 |
| LEF1 | ARHGAP28 | 0.734991 | 3 | 0 | 3 |
| LEF1 | SSH1 | 0.731416 | 3 | 0 | 3 |
| LEF1 | TOR1A | 0.722312 | 3 | 0 | 3 |
| LEF1 | DHRSX | 0.713283 | 3 | 0 | 3 |
| LEF1 | KCNE3 | 0.704459 | 5 | 0 | 5 |
| LEF1 | OAF | 0.701353 | 3 | 0 | 3 |
| LEF1 | OSTC | 0.685353 | 3 | 0 | 3 |
| LEF1 | CELSR2 | 0.675409 | 3 | 0 | 3 |
| LEF1 | ARHGAP9 | 0.671539 | 3 | 0 | 3 |
| LEF1 | SLC30A7 | 0.670423 | 4 | 0 | 3 |
| LEF1 | CHST3 | 0.66608 | 3 | 0 | 3 |
| LEF1 | VMA21 | 0.6606 | 4 | 0 | 3 |
| LEF1 | ESCO2 | 0.657021 | 4 | 0 | 3 |
| LEF1 | POGZ | 0.652873 | 4 | 0 | 4 |
| LEF1 | KRTCAP3 | 0.652708 | 4 | 0 | 4 |
| LEF1 | ZNRF3 | 0.651494 | 4 | 0 | 3 |
| LEF1 | CDCA7 | 0.648463 | 4 | 0 | 3 |
| LEF1 | TMTC4 | 0.643055 | 5 | 0 | 3 |
| LEF1 | KIAA0100 | 0.640979 | 3 | 0 | 3 |
For details and further investigation, click here