| Full name: slingshot protein phosphatase 1 | Alias Symbol: KIAA1298|SSH1L | ||
| Type: protein-coding gene | Cytoband: 12q24.11 | ||
| Entrez ID: 54434 | HGNC ID: HGNC:30579 | Ensembl Gene: ENSG00000084112 | OMIM ID: 606778 |
| Drug and gene relationship at DGIdb | |||
SSH1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance | |
| hsa04810 | Regulation of actin cytoskeleton |
Expression of SSH1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SSH1 | 54434 | 221752_at | 0.2940 | 0.2968 | |
| GSE20347 | SSH1 | 54434 | 221753_at | 0.1746 | 0.1703 | |
| GSE23400 | SSH1 | 54434 | 221753_at | 0.0200 | 0.7492 | |
| GSE26886 | SSH1 | 54434 | 221752_at | -0.2399 | 0.3579 | |
| GSE29001 | SSH1 | 54434 | 221752_at | 0.4066 | 0.0367 | |
| GSE38129 | SSH1 | 54434 | 221752_at | 0.2567 | 0.2597 | |
| GSE45670 | SSH1 | 54434 | 221752_at | 0.0808 | 0.5893 | |
| GSE53622 | SSH1 | 54434 | 51874 | 0.4346 | 0.0000 | |
| GSE53624 | SSH1 | 54434 | 51874 | 0.6299 | 0.0000 | |
| GSE63941 | SSH1 | 54434 | 221752_at | -1.1719 | 0.0338 | |
| GSE77861 | SSH1 | 54434 | 221753_at | 0.3192 | 0.1148 | |
| GSE97050 | SSH1 | 54434 | A_24_P332647 | 0.5922 | 0.1734 | |
| SRP007169 | SSH1 | 54434 | RNAseq | 1.2063 | 0.0013 | |
| SRP008496 | SSH1 | 54434 | RNAseq | 1.2334 | 0.0000 | |
| SRP064894 | SSH1 | 54434 | RNAseq | 0.3552 | 0.1634 | |
| SRP133303 | SSH1 | 54434 | RNAseq | 0.5953 | 0.0031 | |
| SRP159526 | SSH1 | 54434 | RNAseq | 0.0857 | 0.7223 | |
| SRP193095 | SSH1 | 54434 | RNAseq | 0.7245 | 0.0000 | |
| SRP219564 | SSH1 | 54434 | RNAseq | 0.0548 | 0.8344 | |
| TCGA | SSH1 | 54434 | RNAseq | 0.0195 | 0.6689 |
Upregulated datasets: 2; Downregulated datasets: 1.
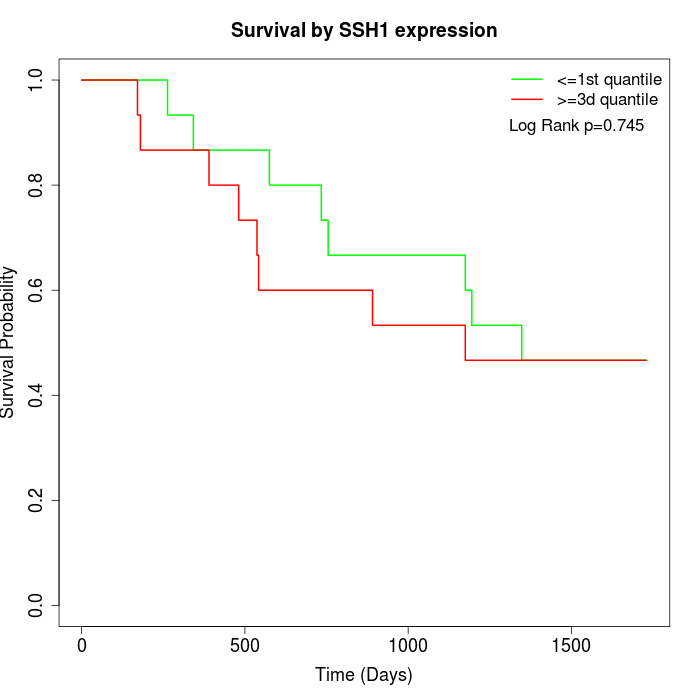
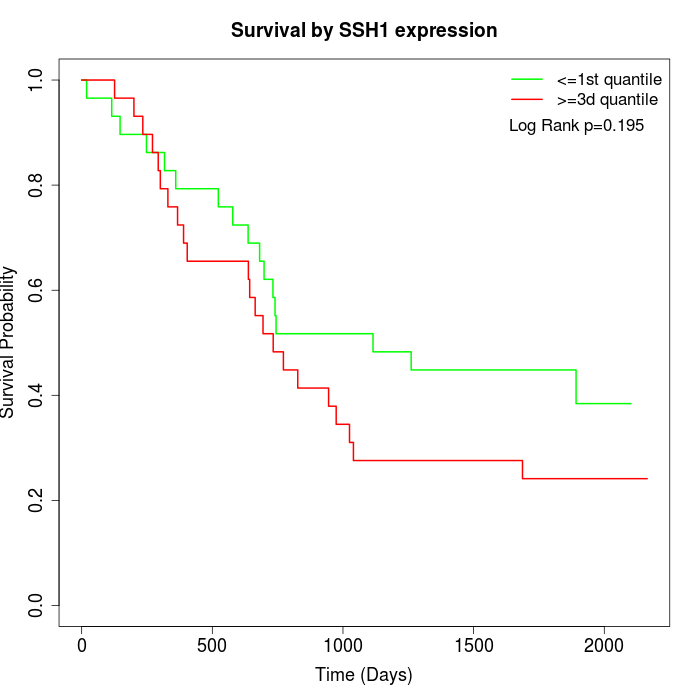
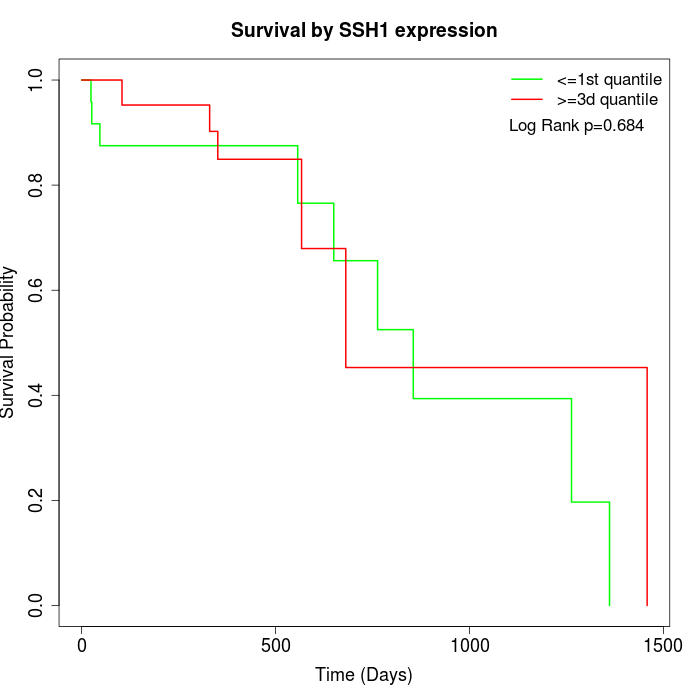
Survival by SSH1 expression:
Note: Click image to view full size file.
Copy number change of SSH1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SSH1 | 54434 | 5 | 3 | 22 | |
| GSE20123 | SSH1 | 54434 | 5 | 4 | 21 | |
| GSE43470 | SSH1 | 54434 | 2 | 1 | 40 | |
| GSE46452 | SSH1 | 54434 | 9 | 1 | 49 | |
| GSE47630 | SSH1 | 54434 | 9 | 1 | 30 | |
| GSE54993 | SSH1 | 54434 | 0 | 5 | 65 | |
| GSE54994 | SSH1 | 54434 | 4 | 2 | 47 | |
| GSE60625 | SSH1 | 54434 | 0 | 0 | 11 | |
| GSE74703 | SSH1 | 54434 | 2 | 0 | 34 | |
| GSE74704 | SSH1 | 54434 | 2 | 2 | 16 | |
| TCGA | SSH1 | 54434 | 21 | 10 | 65 |
Total number of gains: 59; Total number of losses: 29; Total Number of normals: 400.
Somatic mutations of SSH1:
Generating mutation plots.
Highly correlated genes for SSH1:
Showing top 20/691 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SSH1 | TGOLN2 | 0.791755 | 3 | 0 | 3 |
| SSH1 | ARTN | 0.764099 | 3 | 0 | 3 |
| SSH1 | UBE2M | 0.759846 | 3 | 0 | 3 |
| SSH1 | CNRIP1 | 0.755823 | 3 | 0 | 3 |
| SSH1 | FGF7 | 0.747949 | 3 | 0 | 3 |
| SSH1 | NENF | 0.737011 | 3 | 0 | 3 |
| SSH1 | BAP1 | 0.735705 | 3 | 0 | 3 |
| SSH1 | ADAM23 | 0.735494 | 3 | 0 | 3 |
| SSH1 | C20orf194 | 0.733377 | 3 | 0 | 3 |
| SSH1 | LEF1 | 0.731416 | 3 | 0 | 3 |
| SSH1 | RAPGEF6 | 0.728616 | 3 | 0 | 3 |
| SSH1 | TIE1 | 0.726907 | 4 | 0 | 4 |
| SSH1 | CCNK | 0.726003 | 3 | 0 | 3 |
| SSH1 | FCGRT | 0.722702 | 3 | 0 | 3 |
| SSH1 | GNG11 | 0.714929 | 4 | 0 | 4 |
| SSH1 | SPSB2 | 0.712859 | 3 | 0 | 3 |
| SSH1 | DDB1 | 0.711354 | 3 | 0 | 3 |
| SSH1 | MYPOP | 0.710592 | 3 | 0 | 3 |
| SSH1 | ANKRD11 | 0.709026 | 3 | 0 | 3 |
| SSH1 | CD93 | 0.705456 | 4 | 0 | 3 |
For details and further investigation, click here