| Full name: limbic system associated membrane protein | Alias Symbol: LAMP|IGLON3 | ||
| Type: protein-coding gene | Cytoband: 3q13.31 | ||
| Entrez ID: 4045 | HGNC ID: HGNC:6705 | Ensembl Gene: ENSG00000185565 | OMIM ID: 603241 |
| Drug and gene relationship at DGIdb | |||
Expression of LSAMP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LSAMP | 4045 | 229244_at | -0.3712 | 0.3264 | |
| GSE20347 | LSAMP | 4045 | 214460_at | 0.0806 | 0.5734 | |
| GSE23400 | LSAMP | 4045 | 214460_at | -0.0359 | 0.3620 | |
| GSE26886 | LSAMP | 4045 | 229244_at | -0.0928 | 0.5762 | |
| GSE29001 | LSAMP | 4045 | 214460_at | 0.1373 | 0.6193 | |
| GSE38129 | LSAMP | 4045 | 214460_at | 0.0299 | 0.8872 | |
| GSE45670 | LSAMP | 4045 | 229244_at | -0.0482 | 0.6215 | |
| GSE53622 | LSAMP | 4045 | 79412 | 0.1375 | 0.4502 | |
| GSE53624 | LSAMP | 4045 | 79412 | 0.4810 | 0.0119 | |
| GSE63941 | LSAMP | 4045 | 214460_at | -3.5232 | 0.0000 | |
| GSE97050 | LSAMP | 4045 | A_23_P301855 | 0.4179 | 0.2600 | |
| SRP007169 | LSAMP | 4045 | RNAseq | 2.7468 | 0.0006 | |
| SRP008496 | LSAMP | 4045 | RNAseq | 4.5344 | 0.0000 | |
| SRP064894 | LSAMP | 4045 | RNAseq | 0.3149 | 0.4570 | |
| SRP133303 | LSAMP | 4045 | RNAseq | -0.0023 | 0.9918 | |
| SRP159526 | LSAMP | 4045 | RNAseq | 1.4763 | 0.0443 | |
| SRP193095 | LSAMP | 4045 | RNAseq | 1.2127 | 0.0016 | |
| SRP219564 | LSAMP | 4045 | RNAseq | 0.5261 | 0.2867 | |
| TCGA | LSAMP | 4045 | RNAseq | -0.3476 | 0.2187 |
Upregulated datasets: 4; Downregulated datasets: 1.
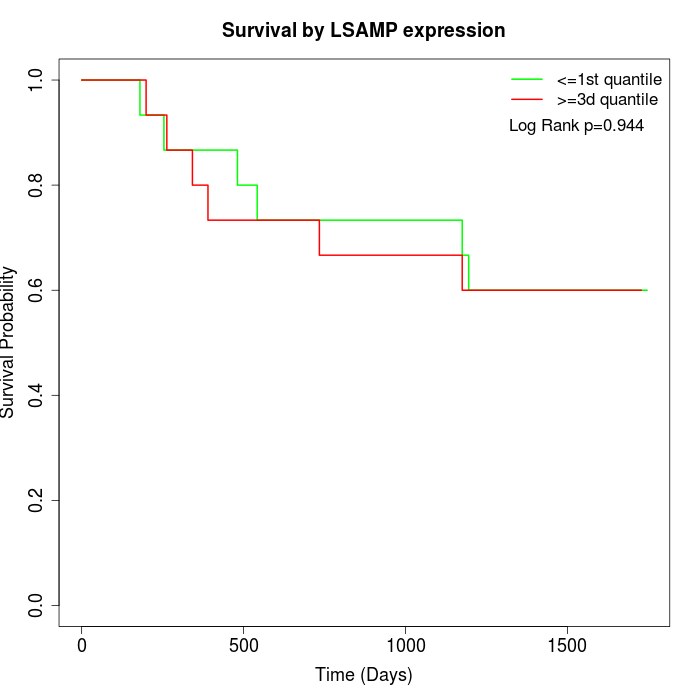
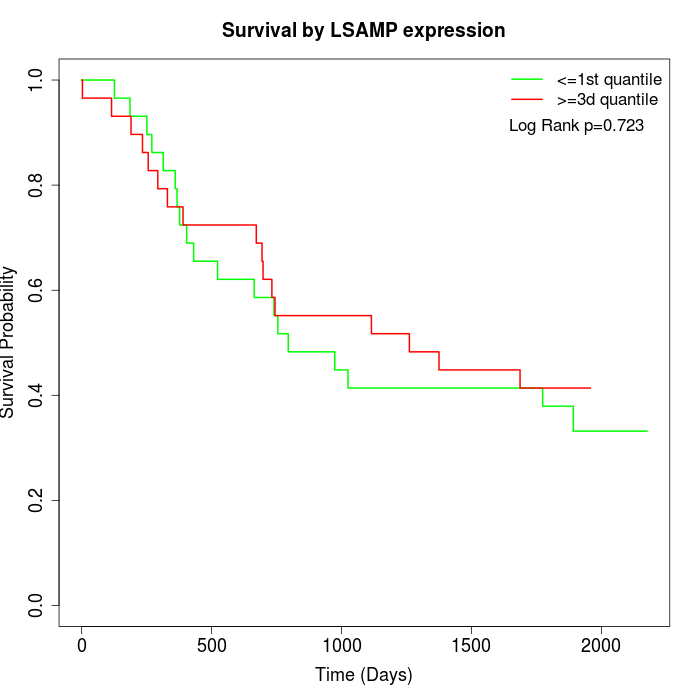
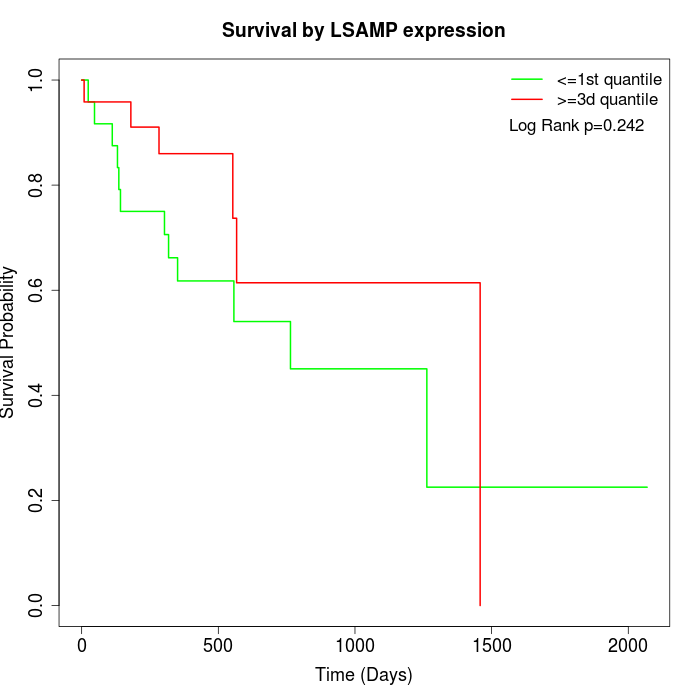
Survival by LSAMP expression:
Note: Click image to view full size file.
Copy number change of LSAMP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LSAMP | 4045 | 17 | 0 | 13 | |
| GSE20123 | LSAMP | 4045 | 17 | 0 | 13 | |
| GSE43470 | LSAMP | 4045 | 21 | 1 | 21 | |
| GSE46452 | LSAMP | 4045 | 14 | 5 | 40 | |
| GSE47630 | LSAMP | 4045 | 15 | 6 | 19 | |
| GSE54993 | LSAMP | 4045 | 2 | 6 | 62 | |
| GSE54994 | LSAMP | 4045 | 29 | 4 | 20 | |
| GSE60625 | LSAMP | 4045 | 0 | 6 | 5 | |
| GSE74703 | LSAMP | 4045 | 17 | 1 | 18 | |
| GSE74704 | LSAMP | 4045 | 13 | 0 | 7 | |
| TCGA | LSAMP | 4045 | 54 | 9 | 33 |
Total number of gains: 199; Total number of losses: 38; Total Number of normals: 251.
Somatic mutations of LSAMP:
Generating mutation plots.
Highly correlated genes for LSAMP:
Showing top 20/107 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LSAMP | FGF3 | 0.719405 | 4 | 0 | 4 |
| LSAMP | CYTL1 | 0.706578 | 3 | 0 | 3 |
| LSAMP | PDE7B | 0.705023 | 3 | 0 | 3 |
| LSAMP | LINC00968 | 0.70268 | 3 | 0 | 3 |
| LSAMP | SCN3A | 0.699426 | 3 | 0 | 3 |
| LSAMP | MAMLD1 | 0.697918 | 3 | 0 | 3 |
| LSAMP | IL33 | 0.689557 | 3 | 0 | 3 |
| LSAMP | VASN | 0.683414 | 3 | 0 | 3 |
| LSAMP | CLEC1A | 0.682808 | 4 | 0 | 4 |
| LSAMP | PCDH18 | 0.675445 | 4 | 0 | 4 |
| LSAMP | VASH2 | 0.669115 | 4 | 0 | 3 |
| LSAMP | QSOX1 | 0.667493 | 4 | 0 | 3 |
| LSAMP | ADAMTS3 | 0.650444 | 5 | 0 | 3 |
| LSAMP | CDH12 | 0.643512 | 4 | 0 | 3 |
| LSAMP | CRABP1 | 0.643392 | 3 | 0 | 3 |
| LSAMP | ADAMTS9-AS2 | 0.639255 | 3 | 0 | 3 |
| LSAMP | NAP1L3 | 0.637977 | 5 | 0 | 3 |
| LSAMP | BEX1 | 0.636472 | 5 | 0 | 3 |
| LSAMP | SNORD114-3 | 0.610403 | 3 | 0 | 3 |
| LSAMP | QPRT | 0.607015 | 5 | 0 | 4 |
For details and further investigation, click here