| Full name: fibroblast growth factor 3 | Alias Symbol: HBGF-3 | ||
| Type: protein-coding gene | Cytoband: 11q13.3 | ||
| Entrez ID: 2248 | HGNC ID: HGNC:3681 | Ensembl Gene: ENSG00000186895 | OMIM ID: 164950 |
| Drug and gene relationship at DGIdb | |||
FGF3 involved pathways:
Expression of FGF3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FGF3 | 2248 | 214571_at | -0.0596 | 0.8118 | |
| GSE20347 | FGF3 | 2248 | 214571_at | 0.2643 | 0.4499 | |
| GSE23400 | FGF3 | 2248 | 214571_at | 0.0315 | 0.8347 | |
| GSE26886 | FGF3 | 2248 | 214571_at | 0.7920 | 0.0363 | |
| GSE29001 | FGF3 | 2248 | 214571_at | 0.3204 | 0.3810 | |
| GSE38129 | FGF3 | 2248 | 214571_at | 0.2277 | 0.3597 | |
| GSE45670 | FGF3 | 2248 | 214571_at | 0.0254 | 0.8306 | |
| GSE63941 | FGF3 | 2248 | 214571_at | -0.0678 | 0.7326 | |
| GSE77861 | FGF3 | 2248 | 214571_at | 0.1295 | 0.6544 | |
| GSE97050 | FGF3 | 2248 | A_33_P3380797 | -0.1741 | 0.4958 | |
| TCGA | FGF3 | 2248 | RNAseq | 2.0943 | 0.2033 |
Upregulated datasets: 0; Downregulated datasets: 0.
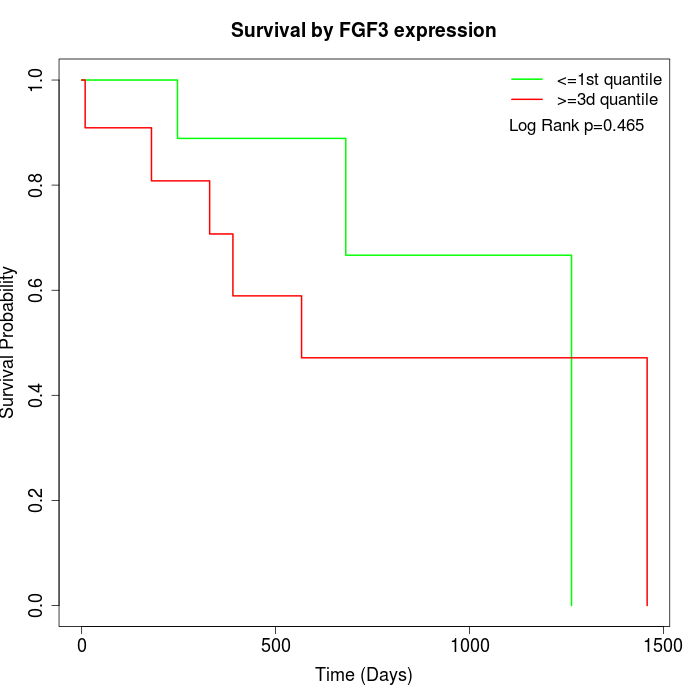
Survival by FGF3 expression:
Note: Click image to view full size file.
Copy number change of FGF3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FGF3 | 2248 | 19 | 1 | 10 | |
| GSE20123 | FGF3 | 2248 | 19 | 1 | 10 | |
| GSE43470 | FGF3 | 2248 | 16 | 2 | 25 | |
| GSE46452 | FGF3 | 2248 | 36 | 1 | 22 | |
| GSE47630 | FGF3 | 2248 | 20 | 1 | 19 | |
| GSE54993 | FGF3 | 2248 | 1 | 25 | 44 | |
| GSE54994 | FGF3 | 2248 | 28 | 3 | 22 | |
| GSE60625 | FGF3 | 2248 | 0 | 8 | 3 | |
| GSE74703 | FGF3 | 2248 | 13 | 1 | 22 | |
| GSE74704 | FGF3 | 2248 | 13 | 0 | 7 | |
| TCGA | FGF3 | 2248 | 63 | 2 | 31 |
Total number of gains: 228; Total number of losses: 45; Total Number of normals: 215.
Somatic mutations of FGF3:
Generating mutation plots.
Highly correlated genes for FGF3:
Showing top 20/117 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FGF3 | MYBPC1 | 0.817862 | 5 | 0 | 5 |
| FGF3 | CNIH3 | 0.799484 | 3 | 0 | 3 |
| FGF3 | NPTX2 | 0.799217 | 6 | 0 | 6 |
| FGF3 | TNFRSF11B | 0.76975 | 6 | 0 | 5 |
| FGF3 | LIFR | 0.744081 | 5 | 0 | 5 |
| FGF3 | LSAMP | 0.719405 | 4 | 0 | 4 |
| FGF3 | CCL2 | 0.714449 | 3 | 0 | 3 |
| FGF3 | PCDH9 | 0.701505 | 3 | 0 | 3 |
| FGF3 | MYCN | 0.698513 | 7 | 0 | 5 |
| FGF3 | PROM1 | 0.694436 | 5 | 0 | 4 |
| FGF3 | SULT1E1 | 0.692757 | 3 | 0 | 3 |
| FGF3 | BEND5 | 0.692303 | 4 | 0 | 3 |
| FGF3 | DBH | 0.691767 | 3 | 0 | 3 |
| FGF3 | HOXB13 | 0.689643 | 5 | 0 | 3 |
| FGF3 | NGFR | 0.688459 | 6 | 0 | 5 |
| FGF3 | WTIP | 0.679254 | 3 | 0 | 3 |
| FGF3 | C11orf71 | 0.679125 | 3 | 0 | 3 |
| FGF3 | SP5 | 0.670391 | 3 | 0 | 3 |
| FGF3 | ARNT2 | 0.670236 | 7 | 0 | 5 |
| FGF3 | IL13RA2 | 0.662319 | 4 | 0 | 3 |
For details and further investigation, click here