| Full name: long intergenic non-protein coding RNA 968 | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 8q12.1 | ||
| Entrez ID: 100507632 | HGNC ID: HGNC:48727 | Ensembl Gene: ENSG00000246430 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LINC00968:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LINC00968 | 100507632 | 243813_at | -0.9161 | 0.0823 | |
| GSE26886 | LINC00968 | 100507632 | 243813_at | 0.0196 | 0.8341 | |
| GSE45670 | LINC00968 | 100507632 | 243813_at | -0.5835 | 0.0013 | |
| GSE53622 | LINC00968 | 100507632 | 62083 | -0.7980 | 0.0000 | |
| GSE53624 | LINC00968 | 100507632 | 62083 | -0.6266 | 0.0016 | |
| GSE63941 | LINC00968 | 100507632 | 243813_at | -4.8749 | 0.0000 | |
| GSE77861 | LINC00968 | 100507632 | 243813_at | -0.0556 | 0.4978 |
Upregulated datasets: 0; Downregulated datasets: 1.
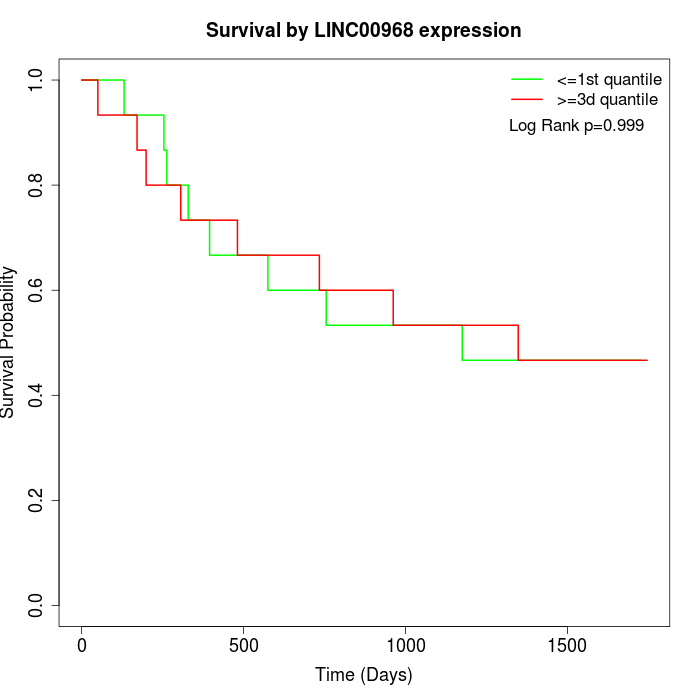
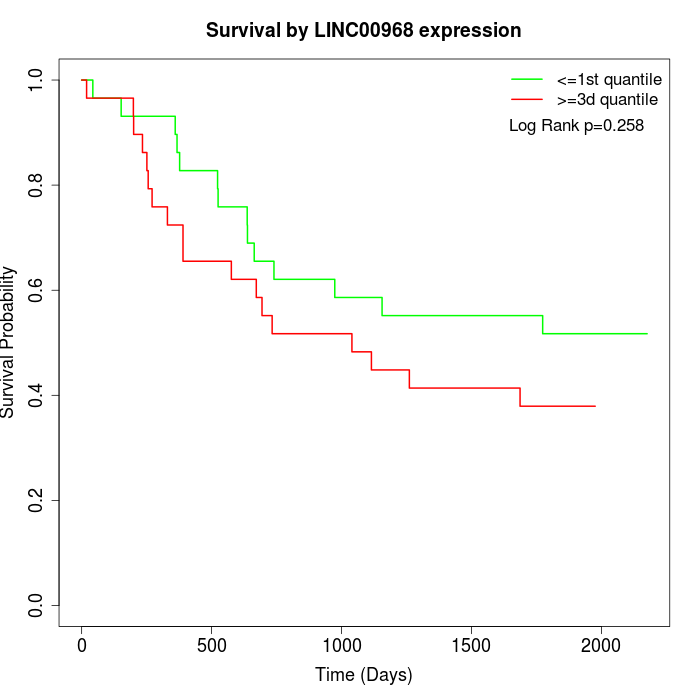
Survival by LINC00968 expression:
Note: Click image to view full size file.
Copy number change of LINC00968:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LINC00968 | 100507632 | 14 | 1 | 15 | |
| GSE20123 | LINC00968 | 100507632 | 14 | 1 | 15 | |
| GSE43470 | LINC00968 | 100507632 | 13 | 3 | 27 | |
| GSE46452 | LINC00968 | 100507632 | 20 | 3 | 36 | |
| GSE47630 | LINC00968 | 100507632 | 23 | 1 | 16 | |
| GSE54993 | LINC00968 | 100507632 | 1 | 17 | 52 | |
| GSE54994 | LINC00968 | 100507632 | 30 | 0 | 23 | |
| GSE60625 | LINC00968 | 100507632 | 0 | 4 | 7 | |
| GSE74703 | LINC00968 | 100507632 | 12 | 2 | 22 | |
| GSE74704 | LINC00968 | 100507632 | 10 | 0 | 10 | |
| TCGA | LINC00968 | 100507632 | 43 | 8 | 45 |
Total number of gains: 180; Total number of losses: 40; Total Number of normals: 268.
Somatic mutations of LINC00968:
Generating mutation plots.
Highly correlated genes for LINC00968:
Showing top 20/345 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LINC00968 | SLIT3 | 0.786085 | 3 | 0 | 3 |
| LINC00968 | INMT | 0.769501 | 3 | 0 | 3 |
| LINC00968 | GSTM5 | 0.759865 | 4 | 0 | 3 |
| LINC00968 | LRRN4CL | 0.744898 | 5 | 0 | 5 |
| LINC00968 | MGARP | 0.732997 | 3 | 0 | 3 |
| LINC00968 | CNGA3 | 0.730987 | 3 | 0 | 3 |
| LINC00968 | FAT4 | 0.730455 | 4 | 0 | 4 |
| LINC00968 | CNTN4 | 0.728061 | 4 | 0 | 3 |
| LINC00968 | ITGBL1 | 0.726436 | 3 | 0 | 3 |
| LINC00968 | SSC5D | 0.718688 | 3 | 0 | 3 |
| LINC00968 | TCF21 | 0.717671 | 5 | 0 | 5 |
| LINC00968 | CPQ | 0.715194 | 4 | 0 | 4 |
| LINC00968 | SMIM10 | 0.712788 | 5 | 0 | 4 |
| LINC00968 | FAM13C | 0.712482 | 3 | 0 | 3 |
| LINC00968 | FBLN5 | 0.709567 | 5 | 0 | 5 |
| LINC00968 | AGTR1 | 0.706701 | 5 | 0 | 4 |
| LINC00968 | IL17D | 0.705666 | 4 | 0 | 3 |
| LINC00968 | LSAMP | 0.70268 | 3 | 0 | 3 |
| LINC00968 | LDB2 | 0.701978 | 4 | 0 | 3 |
| LINC00968 | PRSS35 | 0.700699 | 4 | 0 | 4 |
For details and further investigation, click here