| Full name: microtubule affinity regulating kinase 3 | Alias Symbol: CTAK1|KP78|PAR-1A | ||
| Type: protein-coding gene | Cytoband: 14q32.32-q32.33 | ||
| Entrez ID: 4140 | HGNC ID: HGNC:6897 | Ensembl Gene: ENSG00000075413 | OMIM ID: 602678 |
| Drug and gene relationship at DGIdb | |||
Expression of MARK3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MARK3 | 4140 | 202568_s_at | 0.0900 | 0.9098 | |
| GSE20347 | MARK3 | 4140 | 202568_s_at | 0.0794 | 0.6708 | |
| GSE23400 | MARK3 | 4140 | 202568_s_at | 0.1722 | 0.0385 | |
| GSE26886 | MARK3 | 4140 | 202568_s_at | -0.0431 | 0.8825 | |
| GSE29001 | MARK3 | 4140 | 202569_s_at | 0.1304 | 0.6667 | |
| GSE38129 | MARK3 | 4140 | 202568_s_at | 0.1547 | 0.2407 | |
| GSE45670 | MARK3 | 4140 | 202568_s_at | 0.3534 | 0.0436 | |
| GSE53622 | MARK3 | 4140 | 42192 | 0.2319 | 0.0030 | |
| GSE53624 | MARK3 | 4140 | 42192 | 0.2812 | 0.0000 | |
| GSE63941 | MARK3 | 4140 | 202568_s_at | 0.4196 | 0.2624 | |
| GSE77861 | MARK3 | 4140 | 202569_s_at | 0.2980 | 0.4063 | |
| GSE97050 | MARK3 | 4140 | A_33_P3593546 | 0.2531 | 0.3499 | |
| SRP007169 | MARK3 | 4140 | RNAseq | -0.4398 | 0.2107 | |
| SRP008496 | MARK3 | 4140 | RNAseq | -0.4649 | 0.0252 | |
| SRP064894 | MARK3 | 4140 | RNAseq | 0.0388 | 0.8210 | |
| SRP133303 | MARK3 | 4140 | RNAseq | 0.4543 | 0.0070 | |
| SRP159526 | MARK3 | 4140 | RNAseq | 0.2345 | 0.5289 | |
| SRP193095 | MARK3 | 4140 | RNAseq | 0.2164 | 0.0983 | |
| SRP219564 | MARK3 | 4140 | RNAseq | 0.0415 | 0.9075 | |
| TCGA | MARK3 | 4140 | RNAseq | -0.0078 | 0.8883 |
Upregulated datasets: 0; Downregulated datasets: 0.
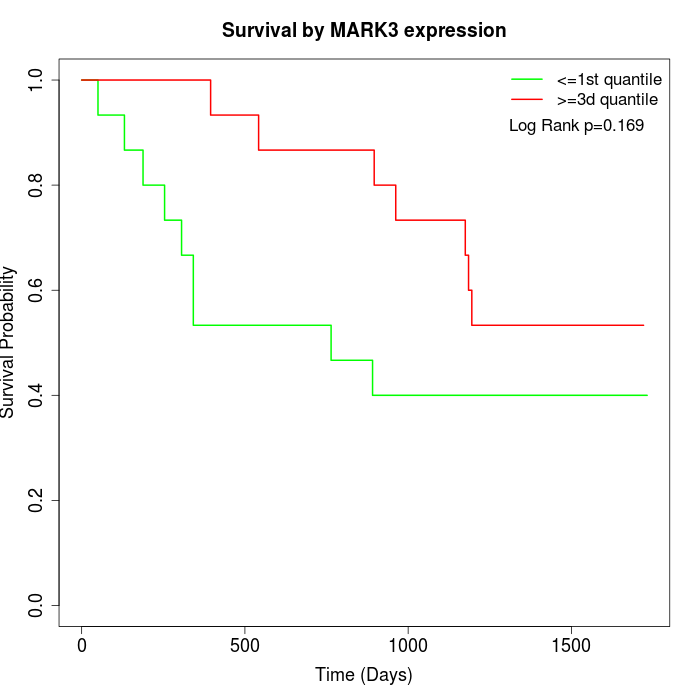
Survival by MARK3 expression:
Note: Click image to view full size file.
Copy number change of MARK3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MARK3 | 4140 | 12 | 3 | 15 | |
| GSE20123 | MARK3 | 4140 | 11 | 2 | 17 | |
| GSE43470 | MARK3 | 4140 | 6 | 4 | 33 | |
| GSE46452 | MARK3 | 4140 | 18 | 4 | 37 | |
| GSE47630 | MARK3 | 4140 | 10 | 8 | 22 | |
| GSE54993 | MARK3 | 4140 | 3 | 8 | 59 | |
| GSE54994 | MARK3 | 4140 | 21 | 4 | 28 | |
| GSE60625 | MARK3 | 4140 | 0 | 2 | 9 | |
| GSE74703 | MARK3 | 4140 | 5 | 3 | 28 | |
| GSE74704 | MARK3 | 4140 | 5 | 2 | 13 | |
| TCGA | MARK3 | 4140 | 33 | 20 | 43 |
Total number of gains: 124; Total number of losses: 60; Total Number of normals: 304.
Somatic mutations of MARK3:
Generating mutation plots.
Highly correlated genes for MARK3:
Showing top 20/412 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MARK3 | TIGD1 | 0.767684 | 3 | 0 | 3 |
| MARK3 | USP40 | 0.746851 | 3 | 0 | 3 |
| MARK3 | RAB1A | 0.723009 | 3 | 0 | 3 |
| MARK3 | SUB1 | 0.711077 | 3 | 0 | 3 |
| MARK3 | MAP3K7 | 0.70759 | 3 | 0 | 3 |
| MARK3 | COX16 | 0.701608 | 4 | 0 | 4 |
| MARK3 | SREK1IP1 | 0.700626 | 3 | 0 | 3 |
| MARK3 | RAB22A | 0.689991 | 5 | 0 | 5 |
| MARK3 | TUBA1B | 0.685004 | 3 | 0 | 3 |
| MARK3 | UBE2E2 | 0.683778 | 3 | 0 | 3 |
| MARK3 | FAM83G | 0.675619 | 4 | 0 | 4 |
| MARK3 | NUDT14 | 0.671603 | 5 | 0 | 4 |
| MARK3 | PPHLN1 | 0.670582 | 3 | 0 | 3 |
| MARK3 | ZFAND2A | 0.669431 | 3 | 0 | 3 |
| MARK3 | LINC00460 | 0.666964 | 3 | 0 | 3 |
| MARK3 | IFI27L2 | 0.666072 | 5 | 0 | 4 |
| MARK3 | EIF2S1 | 0.665739 | 10 | 0 | 10 |
| MARK3 | GOLGA5 | 0.664084 | 9 | 0 | 8 |
| MARK3 | ARPC5L | 0.662261 | 4 | 0 | 3 |
| MARK3 | STON2 | 0.661017 | 8 | 0 | 6 |
For details and further investigation, click here