| Full name: mediator complex subunit 13 | Alias Symbol: KIAA0593|TRAP240 | ||
| Type: protein-coding gene | Cytoband: 17q23.2 | ||
| Entrez ID: 9969 | HGNC ID: HGNC:22474 | Ensembl Gene: ENSG00000108510 | OMIM ID: 603808 |
| Drug and gene relationship at DGIdb | |||
Expression of MED13:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MED13 | 9969 | 201987_at | 0.0998 | 0.6837 | |
| GSE20347 | MED13 | 9969 | 201987_at | 0.0209 | 0.8791 | |
| GSE23400 | MED13 | 9969 | 201987_at | 0.0753 | 0.3198 | |
| GSE26886 | MED13 | 9969 | 201987_at | 0.4162 | 0.0103 | |
| GSE29001 | MED13 | 9969 | 201987_at | 0.1521 | 0.3835 | |
| GSE38129 | MED13 | 9969 | 201987_at | 0.0469 | 0.6808 | |
| GSE45670 | MED13 | 9969 | 201987_at | 0.0327 | 0.8206 | |
| GSE53622 | MED13 | 9969 | 26528 | -0.0685 | 0.1953 | |
| GSE53624 | MED13 | 9969 | 26528 | 0.1127 | 0.0334 | |
| GSE63941 | MED13 | 9969 | 201987_at | -0.9029 | 0.0425 | |
| GSE77861 | MED13 | 9969 | 201987_at | 0.5747 | 0.0599 | |
| GSE97050 | MED13 | 9969 | A_33_P3223759 | -0.1408 | 0.4794 | |
| SRP007169 | MED13 | 9969 | RNAseq | 0.1885 | 0.5198 | |
| SRP008496 | MED13 | 9969 | RNAseq | 0.3030 | 0.0389 | |
| SRP064894 | MED13 | 9969 | RNAseq | -0.1585 | 0.4484 | |
| SRP133303 | MED13 | 9969 | RNAseq | 0.5113 | 0.0020 | |
| SRP159526 | MED13 | 9969 | RNAseq | 0.0431 | 0.7979 | |
| SRP193095 | MED13 | 9969 | RNAseq | -0.0762 | 0.1514 | |
| SRP219564 | MED13 | 9969 | RNAseq | -0.3472 | 0.3198 | |
| TCGA | MED13 | 9969 | RNAseq | 0.0106 | 0.8105 |
Upregulated datasets: 0; Downregulated datasets: 0.
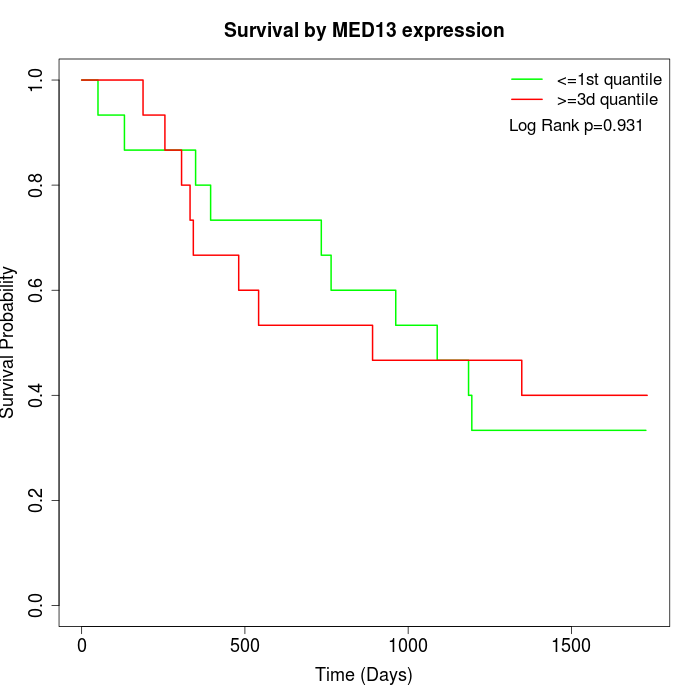
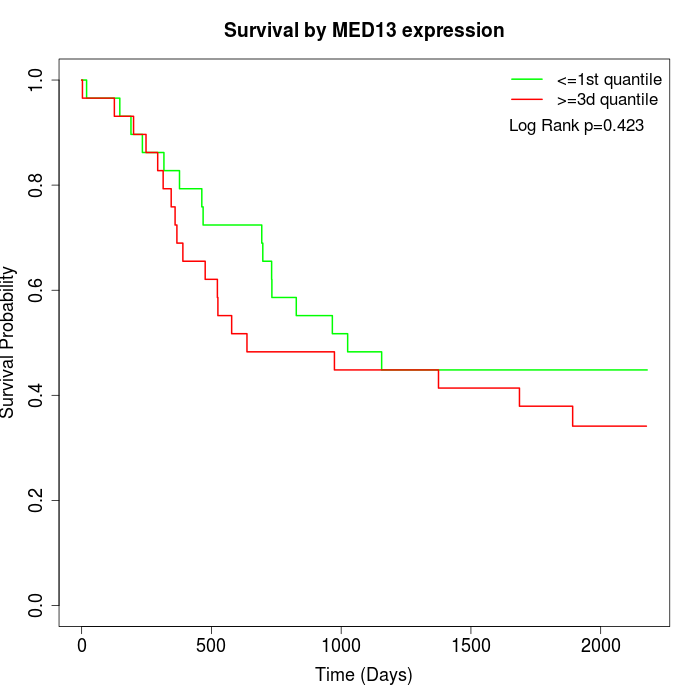
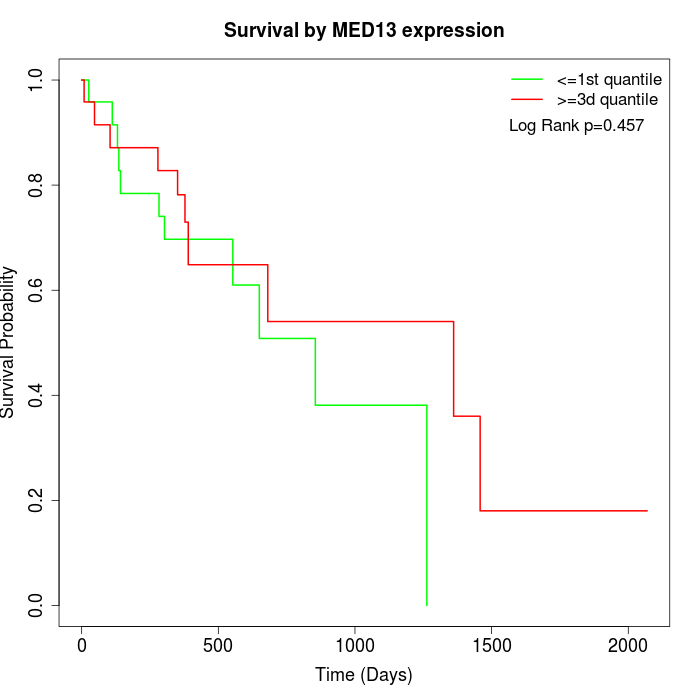
Survival by MED13 expression:
Note: Click image to view full size file.
Copy number change of MED13:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MED13 | 9969 | 4 | 1 | 25 | |
| GSE20123 | MED13 | 9969 | 4 | 1 | 25 | |
| GSE43470 | MED13 | 9969 | 5 | 0 | 38 | |
| GSE46452 | MED13 | 9969 | 31 | 0 | 28 | |
| GSE47630 | MED13 | 9969 | 7 | 1 | 32 | |
| GSE54993 | MED13 | 9969 | 2 | 5 | 63 | |
| GSE54994 | MED13 | 9969 | 9 | 5 | 39 | |
| GSE60625 | MED13 | 9969 | 4 | 0 | 7 | |
| GSE74703 | MED13 | 9969 | 5 | 0 | 31 | |
| GSE74704 | MED13 | 9969 | 3 | 1 | 16 | |
| TCGA | MED13 | 9969 | 31 | 6 | 59 |
Total number of gains: 105; Total number of losses: 20; Total Number of normals: 363.
Somatic mutations of MED13:
Generating mutation plots.
Highly correlated genes for MED13:
Showing top 20/424 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MED13 | U2SURP | 0.758441 | 3 | 0 | 3 |
| MED13 | KIAA0753 | 0.751621 | 3 | 0 | 3 |
| MED13 | ZXDC | 0.751467 | 3 | 0 | 3 |
| MED13 | GLS | 0.750039 | 3 | 0 | 3 |
| MED13 | SPSB2 | 0.741721 | 3 | 0 | 3 |
| MED13 | CUL7 | 0.740305 | 4 | 0 | 4 |
| MED13 | MEA1 | 0.736837 | 3 | 0 | 3 |
| MED13 | CHERP | 0.735438 | 4 | 0 | 4 |
| MED13 | RRAS2 | 0.7341 | 3 | 0 | 3 |
| MED13 | DNAJC13 | 0.733978 | 3 | 0 | 3 |
| MED13 | ZBTB39 | 0.73155 | 4 | 0 | 4 |
| MED13 | GNA13 | 0.730194 | 5 | 0 | 5 |
| MED13 | ZNF260 | 0.727835 | 4 | 0 | 3 |
| MED13 | NVL | 0.72619 | 3 | 0 | 3 |
| MED13 | DHX40 | 0.726002 | 4 | 0 | 4 |
| MED13 | ZNF397 | 0.724959 | 3 | 0 | 3 |
| MED13 | ITGAV | 0.723364 | 3 | 0 | 3 |
| MED13 | NOL9 | 0.723081 | 3 | 0 | 3 |
| MED13 | SNAPC4 | 0.722937 | 3 | 0 | 3 |
| MED13 | PIK3R3 | 0.722188 | 3 | 0 | 3 |
For details and further investigation, click here