| Full name: integrin subunit alpha V | Alias Symbol: CD51 | ||
| Type: protein-coding gene | Cytoband: 2q32.1 | ||
| Entrez ID: 3685 | HGNC ID: HGNC:6150 | Ensembl Gene: ENSG00000138448 | OMIM ID: 193210 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ITGAV involved pathways:
Expression of ITGAV:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ITGAV | 3685 | 202351_at | 0.7444 | 0.1779 | |
| GSE20347 | ITGAV | 3685 | 202351_at | 1.0568 | 0.0001 | |
| GSE23400 | ITGAV | 3685 | 202351_at | 1.0582 | 0.0000 | |
| GSE26886 | ITGAV | 3685 | 202351_at | 1.3835 | 0.0000 | |
| GSE29001 | ITGAV | 3685 | 202351_at | 1.1085 | 0.0048 | |
| GSE38129 | ITGAV | 3685 | 202351_at | 1.0151 | 0.0000 | |
| GSE45670 | ITGAV | 3685 | 202351_at | 0.5132 | 0.0764 | |
| GSE53622 | ITGAV | 3685 | 12561 | 1.1745 | 0.0000 | |
| GSE53624 | ITGAV | 3685 | 12561 | 1.1844 | 0.0000 | |
| GSE63941 | ITGAV | 3685 | 202351_at | -2.8106 | 0.1652 | |
| GSE77861 | ITGAV | 3685 | 202351_at | 1.4187 | 0.0038 | |
| GSE97050 | ITGAV | 3685 | A_23_P50907 | 0.6548 | 0.0970 | |
| SRP007169 | ITGAV | 3685 | RNAseq | 2.3349 | 0.0000 | |
| SRP008496 | ITGAV | 3685 | RNAseq | 2.3671 | 0.0000 | |
| SRP064894 | ITGAV | 3685 | RNAseq | 1.5207 | 0.0000 | |
| SRP133303 | ITGAV | 3685 | RNAseq | 1.8427 | 0.0000 | |
| SRP159526 | ITGAV | 3685 | RNAseq | 0.8091 | 0.0000 | |
| SRP193095 | ITGAV | 3685 | RNAseq | 1.1108 | 0.0000 | |
| SRP219564 | ITGAV | 3685 | RNAseq | 0.3106 | 0.5268 | |
| TCGA | ITGAV | 3685 | RNAseq | 0.0754 | 0.2703 |
Upregulated datasets: 13; Downregulated datasets: 0.
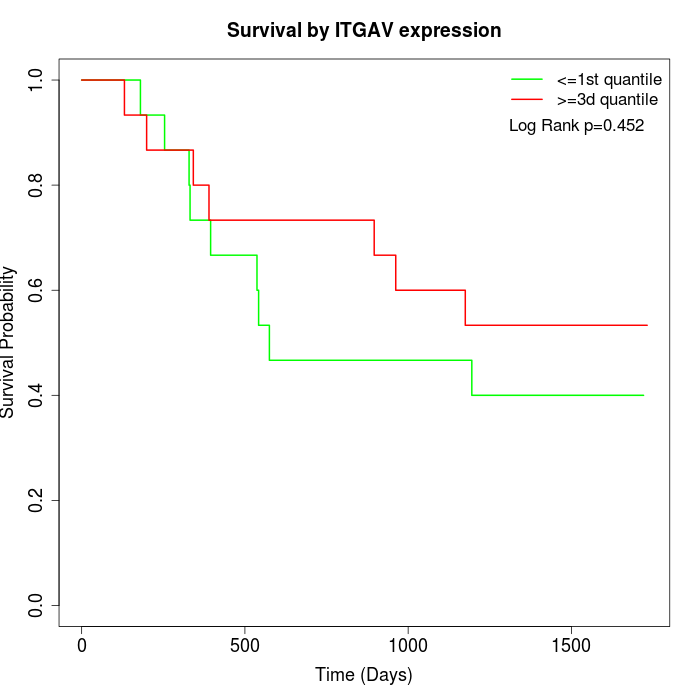
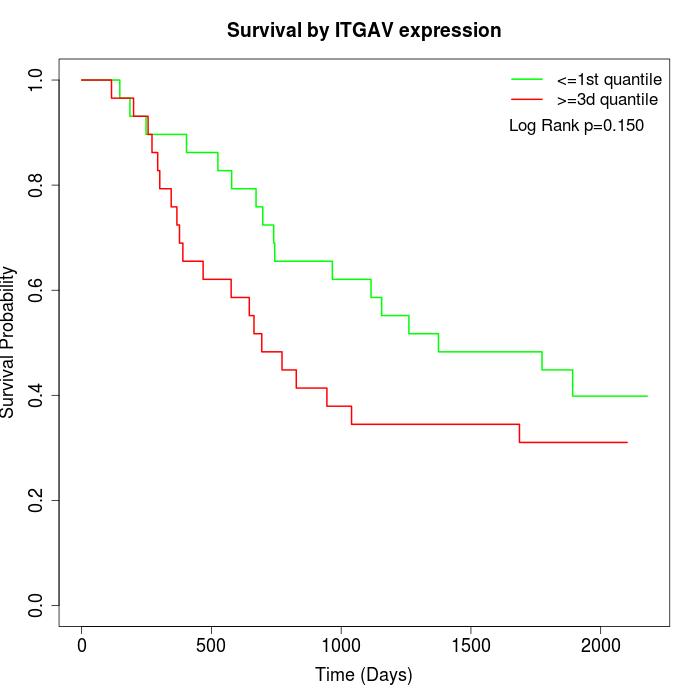
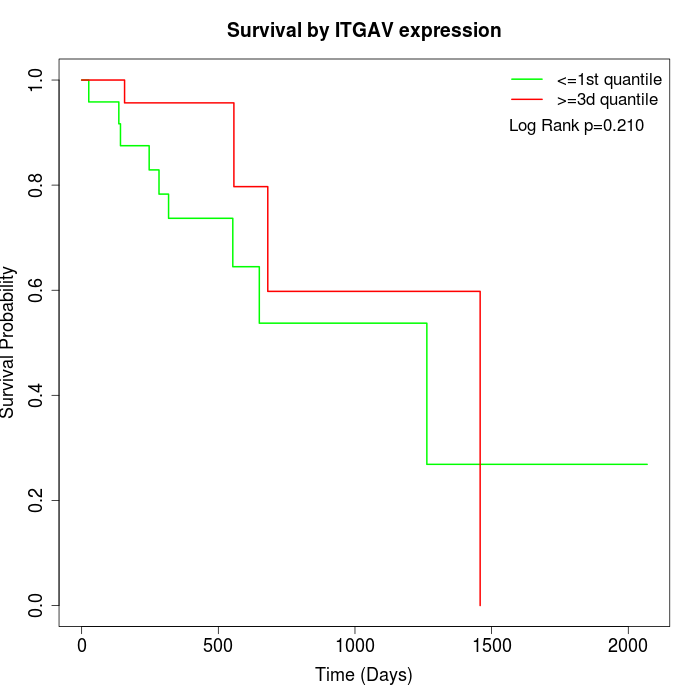
Survival by ITGAV expression:
Note: Click image to view full size file.
Copy number change of ITGAV:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ITGAV | 3685 | 6 | 1 | 23 | |
| GSE20123 | ITGAV | 3685 | 7 | 1 | 22 | |
| GSE43470 | ITGAV | 3685 | 3 | 1 | 39 | |
| GSE46452 | ITGAV | 3685 | 1 | 4 | 54 | |
| GSE47630 | ITGAV | 3685 | 4 | 5 | 31 | |
| GSE54993 | ITGAV | 3685 | 0 | 5 | 65 | |
| GSE54994 | ITGAV | 3685 | 12 | 6 | 35 | |
| GSE60625 | ITGAV | 3685 | 0 | 3 | 8 | |
| GSE74703 | ITGAV | 3685 | 2 | 1 | 33 | |
| GSE74704 | ITGAV | 3685 | 3 | 1 | 16 | |
| TCGA | ITGAV | 3685 | 24 | 9 | 63 |
Total number of gains: 62; Total number of losses: 37; Total Number of normals: 389.
Somatic mutations of ITGAV:
Generating mutation plots.
Highly correlated genes for ITGAV:
Showing top 20/1908 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ITGAV | HAS3 | 0.800788 | 3 | 0 | 3 |
| ITGAV | TMTC1 | 0.792776 | 3 | 0 | 3 |
| ITGAV | INHBA | 0.786997 | 10 | 0 | 10 |
| ITGAV | LRP12 | 0.776865 | 11 | 0 | 11 |
| ITGAV | CEP120 | 0.776239 | 3 | 0 | 3 |
| ITGAV | ACVR1 | 0.775518 | 13 | 0 | 12 |
| ITGAV | SLC39A14 | 0.770329 | 12 | 0 | 12 |
| ITGAV | ENAH | 0.76612 | 12 | 0 | 11 |
| ITGAV | COL5A2 | 0.761661 | 12 | 0 | 11 |
| ITGAV | SLC39A3 | 0.75609 | 3 | 0 | 3 |
| ITGAV | TGFBI | 0.755544 | 13 | 0 | 12 |
| ITGAV | MFAP2 | 0.751614 | 11 | 0 | 11 |
| ITGAV | WDR54 | 0.750181 | 7 | 0 | 7 |
| ITGAV | XPR1 | 0.749557 | 7 | 0 | 7 |
| ITGAV | PRNP | 0.748424 | 12 | 0 | 11 |
| ITGAV | SDHAF2 | 0.74768 | 3 | 0 | 3 |
| ITGAV | STAT2 | 0.747508 | 4 | 0 | 3 |
| ITGAV | CHIC1 | 0.746166 | 3 | 0 | 3 |
| ITGAV | CD276 | 0.745856 | 4 | 0 | 4 |
| ITGAV | A4GALT | 0.740652 | 3 | 0 | 3 |
For details and further investigation, click here