| Full name: RAS related 2 | Alias Symbol: TC21 | ||
| Type: protein-coding gene | Cytoband: 11p15.2 | ||
| Entrez ID: 22800 | HGNC ID: HGNC:17271 | Ensembl Gene: ENSG00000133818 | OMIM ID: 600098 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
RRAS2 involved pathways:
Expression of RRAS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RRAS2 | 22800 | 212589_at | 0.7270 | 0.0770 | |
| GSE20347 | RRAS2 | 22800 | 212589_at | 0.6048 | 0.0425 | |
| GSE23400 | RRAS2 | 22800 | 212589_at | 0.8592 | 0.0000 | |
| GSE26886 | RRAS2 | 22800 | 212589_at | 1.0969 | 0.0001 | |
| GSE29001 | RRAS2 | 22800 | 212589_at | 1.1269 | 0.0002 | |
| GSE38129 | RRAS2 | 22800 | 212589_at | 0.4832 | 0.0185 | |
| GSE45670 | RRAS2 | 22800 | 212589_at | 0.5085 | 0.0579 | |
| GSE63941 | RRAS2 | 22800 | 212589_at | -1.2634 | 0.0128 | |
| GSE77861 | RRAS2 | 22800 | 212589_at | 1.4563 | 0.0166 | |
| GSE97050 | RRAS2 | 22800 | A_33_P3297245 | 0.3287 | 0.3693 | |
| SRP007169 | RRAS2 | 22800 | RNAseq | 0.5549 | 0.2692 | |
| SRP008496 | RRAS2 | 22800 | RNAseq | 0.8048 | 0.0035 | |
| SRP064894 | RRAS2 | 22800 | RNAseq | 0.3032 | 0.1105 | |
| SRP133303 | RRAS2 | 22800 | RNAseq | 1.5781 | 0.0000 | |
| SRP159526 | RRAS2 | 22800 | RNAseq | 0.3921 | 0.3339 | |
| SRP193095 | RRAS2 | 22800 | RNAseq | 0.6367 | 0.0001 | |
| SRP219564 | RRAS2 | 22800 | RNAseq | 0.4219 | 0.2127 | |
| TCGA | RRAS2 | 22800 | RNAseq | 0.1755 | 0.0044 |
Upregulated datasets: 4; Downregulated datasets: 1.
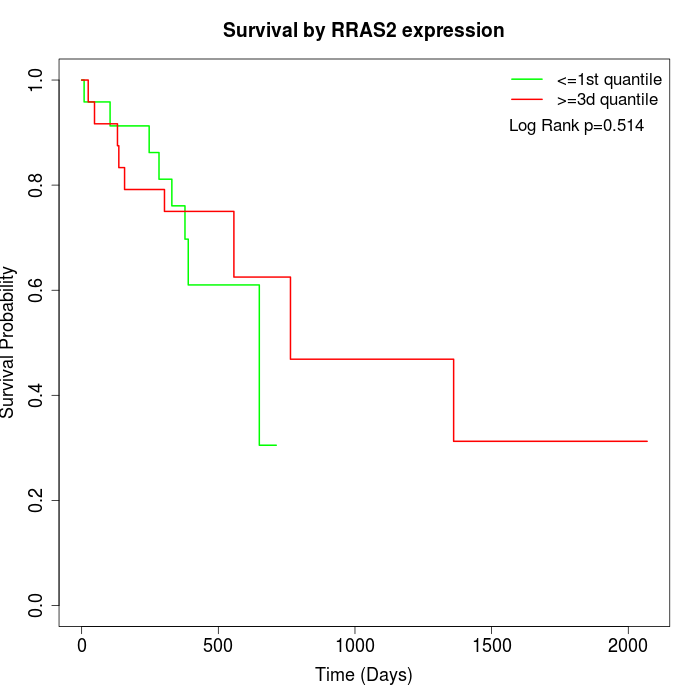
Survival by RRAS2 expression:
Note: Click image to view full size file.
Copy number change of RRAS2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RRAS2 | 22800 | 1 | 10 | 19 | |
| GSE20123 | RRAS2 | 22800 | 1 | 10 | 19 | |
| GSE43470 | RRAS2 | 22800 | 1 | 5 | 37 | |
| GSE46452 | RRAS2 | 22800 | 7 | 5 | 47 | |
| GSE47630 | RRAS2 | 22800 | 3 | 11 | 26 | |
| GSE54993 | RRAS2 | 22800 | 3 | 1 | 66 | |
| GSE54994 | RRAS2 | 22800 | 4 | 12 | 37 | |
| GSE60625 | RRAS2 | 22800 | 0 | 0 | 11 | |
| GSE74703 | RRAS2 | 22800 | 1 | 3 | 32 | |
| GSE74704 | RRAS2 | 22800 | 0 | 9 | 11 | |
| TCGA | RRAS2 | 22800 | 12 | 31 | 53 |
Total number of gains: 33; Total number of losses: 97; Total Number of normals: 358.
Somatic mutations of RRAS2:
Generating mutation plots.
Highly correlated genes for RRAS2:
Showing top 20/1185 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RRAS2 | SOCS7 | 0.816707 | 3 | 0 | 3 |
| RRAS2 | TRIM35 | 0.796517 | 3 | 0 | 3 |
| RRAS2 | PLEKHM3 | 0.780474 | 3 | 0 | 3 |
| RRAS2 | A4GALT | 0.772843 | 3 | 0 | 3 |
| RRAS2 | RBM22 | 0.771551 | 3 | 0 | 3 |
| RRAS2 | RBM18 | 0.768978 | 3 | 0 | 3 |
| RRAS2 | RNASEH2C | 0.768725 | 3 | 0 | 3 |
| RRAS2 | NOSIP | 0.767506 | 3 | 0 | 3 |
| RRAS2 | NAPEPLD | 0.760948 | 3 | 0 | 3 |
| RRAS2 | IFFO2 | 0.760238 | 3 | 0 | 3 |
| RRAS2 | PRTFDC1 | 0.760193 | 3 | 0 | 3 |
| RRAS2 | ARL13B | 0.758747 | 4 | 0 | 3 |
| RRAS2 | YWHAG | 0.758023 | 5 | 0 | 5 |
| RRAS2 | KLC1 | 0.75681 | 3 | 0 | 3 |
| RRAS2 | MASTL | 0.753817 | 3 | 0 | 3 |
| RRAS2 | TNPO1 | 0.751608 | 3 | 0 | 3 |
| RRAS2 | RRM2B | 0.749412 | 3 | 0 | 3 |
| RRAS2 | CTR9 | 0.74824 | 3 | 0 | 3 |
| RRAS2 | BHLHE41 | 0.747592 | 3 | 0 | 3 |
| RRAS2 | ZNF354A | 0.742538 | 4 | 0 | 4 |
For details and further investigation, click here