| Full name: mediator complex subunit 22 | Alias Symbol: Med24|SRB6 | ||
| Type: protein-coding gene | Cytoband: 9q34.2 | ||
| Entrez ID: 6837 | HGNC ID: HGNC:11477 | Ensembl Gene: ENSG00000148297 | OMIM ID: 185641 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of MED22:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MED22 | 6837 | 206593_s_at | 0.2449 | 0.5621 | |
| GSE20347 | MED22 | 6837 | 206593_s_at | 0.2026 | 0.0937 | |
| GSE23400 | MED22 | 6837 | 206593_s_at | 0.1111 | 0.0873 | |
| GSE26886 | MED22 | 6837 | 206593_s_at | 0.8788 | 0.0000 | |
| GSE29001 | MED22 | 6837 | 206593_s_at | 0.4173 | 0.0276 | |
| GSE38129 | MED22 | 6837 | 206593_s_at | 0.1234 | 0.3920 | |
| GSE45670 | MED22 | 6837 | 206593_s_at | 0.0760 | 0.5479 | |
| GSE53622 | MED22 | 6837 | 136098 | -0.3353 | 0.0000 | |
| GSE53624 | MED22 | 6837 | 136098 | -0.9126 | 0.0000 | |
| GSE63941 | MED22 | 6837 | 206593_s_at | 0.2166 | 0.5786 | |
| GSE77861 | MED22 | 6837 | 206593_s_at | 0.3555 | 0.0793 | |
| GSE97050 | MED22 | 6837 | A_33_P3350227 | -0.0837 | 0.6706 | |
| SRP007169 | MED22 | 6837 | RNAseq | 0.5978 | 0.2122 | |
| SRP008496 | MED22 | 6837 | RNAseq | 0.7770 | 0.0051 | |
| SRP064894 | MED22 | 6837 | RNAseq | 0.6176 | 0.0047 | |
| SRP133303 | MED22 | 6837 | RNAseq | 0.1049 | 0.5155 | |
| SRP159526 | MED22 | 6837 | RNAseq | 0.3088 | 0.3008 | |
| SRP193095 | MED22 | 6837 | RNAseq | 0.4047 | 0.0000 | |
| SRP219564 | MED22 | 6837 | RNAseq | -0.0862 | 0.7983 | |
| TCGA | MED22 | 6837 | RNAseq | -0.0111 | 0.8414 |
Upregulated datasets: 0; Downregulated datasets: 0.
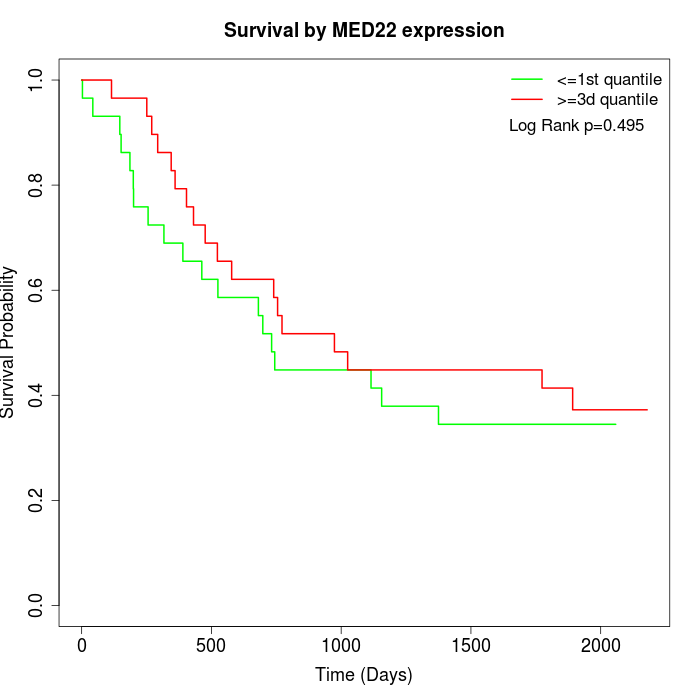
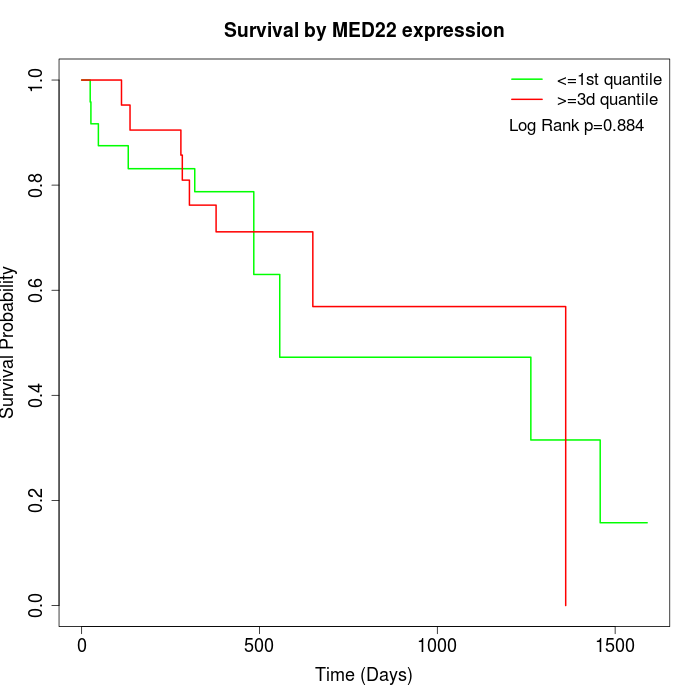
Survival by MED22 expression:
Note: Click image to view full size file.
Copy number change of MED22:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MED22 | 6837 | 5 | 7 | 18 | |
| GSE20123 | MED22 | 6837 | 5 | 7 | 18 | |
| GSE43470 | MED22 | 6837 | 4 | 7 | 32 | |
| GSE46452 | MED22 | 6837 | 6 | 13 | 40 | |
| GSE47630 | MED22 | 6837 | 4 | 15 | 21 | |
| GSE54993 | MED22 | 6837 | 3 | 3 | 64 | |
| GSE54994 | MED22 | 6837 | 12 | 9 | 32 | |
| GSE60625 | MED22 | 6837 | 0 | 0 | 11 | |
| GSE74703 | MED22 | 6837 | 4 | 5 | 27 | |
| GSE74704 | MED22 | 6837 | 3 | 5 | 12 | |
| TCGA | MED22 | 6837 | 28 | 26 | 42 |
Total number of gains: 74; Total number of losses: 97; Total Number of normals: 317.
Somatic mutations of MED22:
Generating mutation plots.
Highly correlated genes for MED22:
Showing top 20/503 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MED22 | DHRSX | 0.830179 | 3 | 0 | 3 |
| MED22 | TAF1C | 0.816774 | 3 | 0 | 3 |
| MED22 | SSBP4 | 0.8081 | 4 | 0 | 4 |
| MED22 | CASC3 | 0.790967 | 3 | 0 | 3 |
| MED22 | ATRNL1 | 0.779676 | 4 | 0 | 4 |
| MED22 | TRIM39 | 0.772279 | 3 | 0 | 3 |
| MED22 | NR1H2 | 0.754144 | 4 | 0 | 4 |
| MED22 | CCDC124 | 0.744015 | 4 | 0 | 4 |
| MED22 | EEF2 | 0.742312 | 4 | 0 | 3 |
| MED22 | DNAAF2 | 0.737796 | 3 | 0 | 3 |
| MED22 | ALKBH6 | 0.734444 | 3 | 0 | 3 |
| MED22 | KRTCAP2 | 0.732224 | 4 | 0 | 3 |
| MED22 | CAMKK1 | 0.732074 | 3 | 0 | 3 |
| MED22 | ZNF830 | 0.729879 | 3 | 0 | 3 |
| MED22 | PRSS36 | 0.727874 | 3 | 0 | 3 |
| MED22 | C20orf96 | 0.727329 | 3 | 0 | 3 |
| MED22 | PPP5C | 0.726568 | 4 | 0 | 3 |
| MED22 | LACTBL1 | 0.726248 | 3 | 0 | 3 |
| MED22 | RAB11FIP5 | 0.725313 | 3 | 0 | 3 |
| MED22 | DAZAP1 | 0.724075 | 3 | 0 | 3 |
For details and further investigation, click here