| Full name: tripartite motif containing 39 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 6p22.1 | ||
| Entrez ID: 56658 | HGNC ID: HGNC:10065 | Ensembl Gene: ENSG00000204599 | OMIM ID: 605700 |
| Drug and gene relationship at DGIdb | |||
Expression of TRIM39:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | TRIM39 | 56658 | 110805 | -0.8730 | 0.0000 | |
| GSE53624 | TRIM39 | 56658 | 110805 | -0.5191 | 0.0000 | |
| GSE97050 | TRIM39 | 56658 | A_33_P3306217 | -0.1322 | 0.4715 | |
| SRP007169 | TRIM39 | 56658 | RNAseq | -0.9650 | 0.0185 | |
| SRP008496 | TRIM39 | 56658 | RNAseq | -0.9591 | 0.0002 | |
| SRP064894 | TRIM39 | 56658 | RNAseq | -0.4299 | 0.0018 | |
| SRP133303 | TRIM39 | 56658 | RNAseq | -0.2746 | 0.0645 | |
| SRP159526 | TRIM39 | 56658 | RNAseq | -0.4631 | 0.1095 | |
| SRP193095 | TRIM39 | 56658 | RNAseq | -0.4232 | 0.0000 | |
| SRP219564 | TRIM39 | 56658 | RNAseq | -0.4244 | 0.1491 | |
| TCGA | TRIM39 | 56658 | RNAseq | -0.2025 | 0.0004 |
Upregulated datasets: 0; Downregulated datasets: 0.
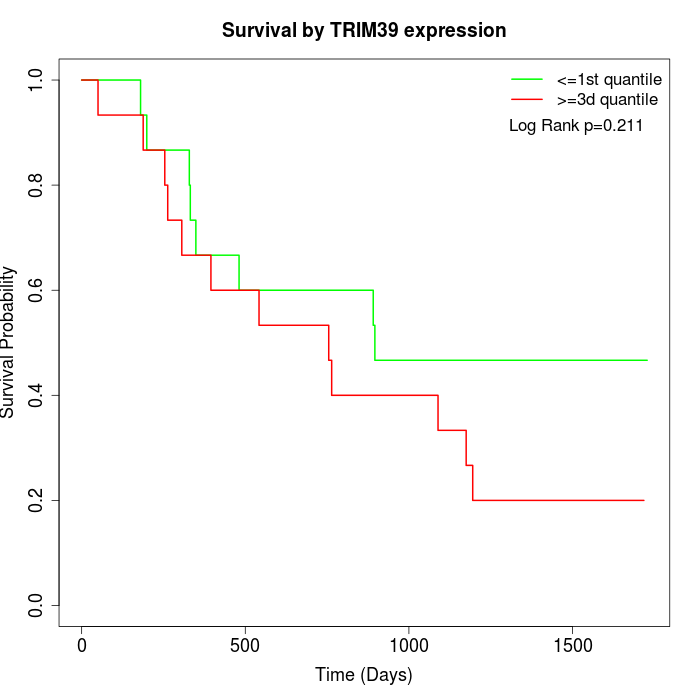
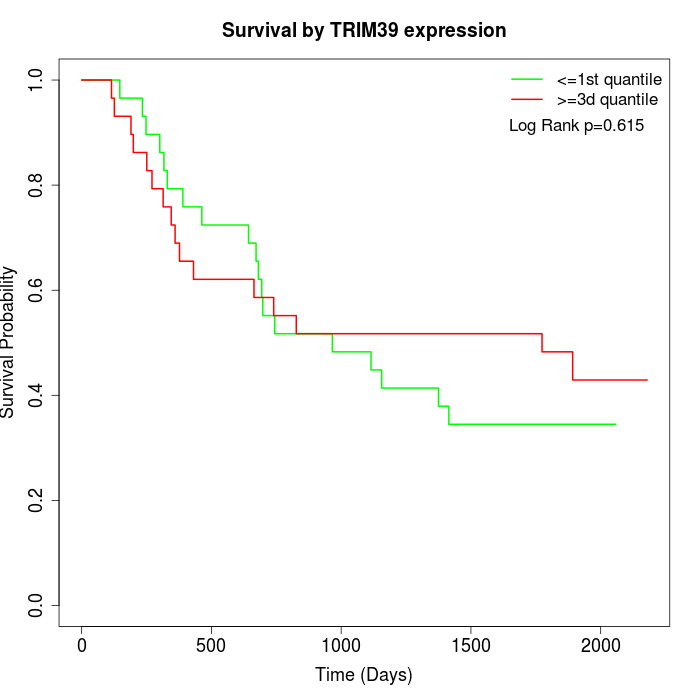
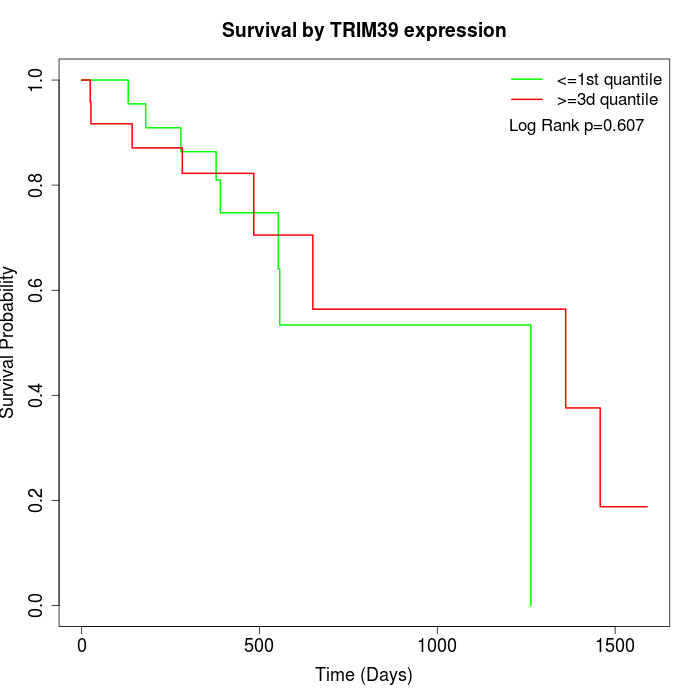
Survival by TRIM39 expression:
Note: Click image to view full size file.
Copy number change of TRIM39:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM39 | 56658 | 5 | 1 | 24 | |
| GSE20123 | TRIM39 | 56658 | 5 | 1 | 24 | |
| GSE43470 | TRIM39 | 56658 | 5 | 1 | 37 | |
| GSE46452 | TRIM39 | 56658 | 1 | 10 | 48 | |
| GSE47630 | TRIM39 | 56658 | 7 | 3 | 30 | |
| GSE54993 | TRIM39 | 56658 | 2 | 1 | 67 | |
| GSE54994 | TRIM39 | 56658 | 11 | 4 | 38 | |
| GSE60625 | TRIM39 | 56658 | 0 | 1 | 10 | |
| GSE74703 | TRIM39 | 56658 | 5 | 0 | 31 | |
| GSE74704 | TRIM39 | 56658 | 2 | 0 | 18 | |
| TCGA | TRIM39 | 56658 | 17 | 16 | 63 |
Total number of gains: 60; Total number of losses: 38; Total Number of normals: 390.
Somatic mutations of TRIM39:
Generating mutation plots.
Highly correlated genes for TRIM39:
Showing top 20/53 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM39 | ATRNL1 | 0.790761 | 3 | 0 | 3 |
| TRIM39 | MED22 | 0.772279 | 3 | 0 | 3 |
| TRIM39 | CUEDC1 | 0.765159 | 3 | 0 | 3 |
| TRIM39 | ADAD2 | 0.745302 | 3 | 0 | 3 |
| TRIM39 | DIP2A | 0.745043 | 3 | 0 | 3 |
| TRIM39 | NEUROG3 | 0.71523 | 3 | 0 | 3 |
| TRIM39 | LACTBL1 | 0.705983 | 3 | 0 | 3 |
| TRIM39 | CDX1 | 0.704902 | 3 | 0 | 3 |
| TRIM39 | CPEB4 | 0.697658 | 3 | 0 | 3 |
| TRIM39 | RNF141 | 0.692325 | 3 | 0 | 3 |
| TRIM39 | SLK | 0.69068 | 3 | 0 | 3 |
| TRIM39 | CLIP1 | 0.690369 | 3 | 0 | 3 |
| TRIM39 | LHX3 | 0.69007 | 3 | 0 | 3 |
| TRIM39 | SLC38A7 | 0.677503 | 3 | 0 | 3 |
| TRIM39 | CRHR1 | 0.675009 | 3 | 0 | 3 |
| TRIM39 | DNAAF2 | 0.673741 | 3 | 0 | 3 |
| TRIM39 | MPP7 | 0.670209 | 3 | 0 | 3 |
| TRIM39 | NDUFC1 | 0.664261 | 3 | 0 | 3 |
| TRIM39 | TF | 0.663722 | 3 | 0 | 3 |
| TRIM39 | KAT2B | 0.663174 | 3 | 0 | 3 |
For details and further investigation, click here