| Full name: serine protease 36 | Alias Symbol: FLJ90661 | ||
| Type: protein-coding gene | Cytoband: 16p11.2 | ||
| Entrez ID: 146547 | HGNC ID: HGNC:26906 | Ensembl Gene: ENSG00000178226 | OMIM ID: 610560 |
| Drug and gene relationship at DGIdb | |||
Expression of PRSS36:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRSS36 | 146547 | 1552555_at | 0.1905 | 0.4819 | |
| GSE26886 | PRSS36 | 146547 | 1552555_at | 0.1897 | 0.2004 | |
| GSE45670 | PRSS36 | 146547 | 1552555_at | 0.1876 | 0.0946 | |
| GSE53622 | PRSS36 | 146547 | 33259 | -0.0940 | 0.1628 | |
| GSE53624 | PRSS36 | 146547 | 33259 | -0.2519 | 0.1268 | |
| GSE63941 | PRSS36 | 146547 | 1552555_at | 0.3691 | 0.0788 | |
| GSE77861 | PRSS36 | 146547 | 1552555_at | 0.0968 | 0.5551 | |
| GSE97050 | PRSS36 | 146547 | A_24_P412734 | -0.0305 | 0.9259 | |
| SRP133303 | PRSS36 | 146547 | RNAseq | 0.0752 | 0.8015 | |
| SRP159526 | PRSS36 | 146547 | RNAseq | 0.0794 | 0.8486 | |
| SRP219564 | PRSS36 | 146547 | RNAseq | 0.5595 | 0.1357 | |
| TCGA | PRSS36 | 146547 | RNAseq | -0.2519 | 0.1301 |
Upregulated datasets: 0; Downregulated datasets: 0.
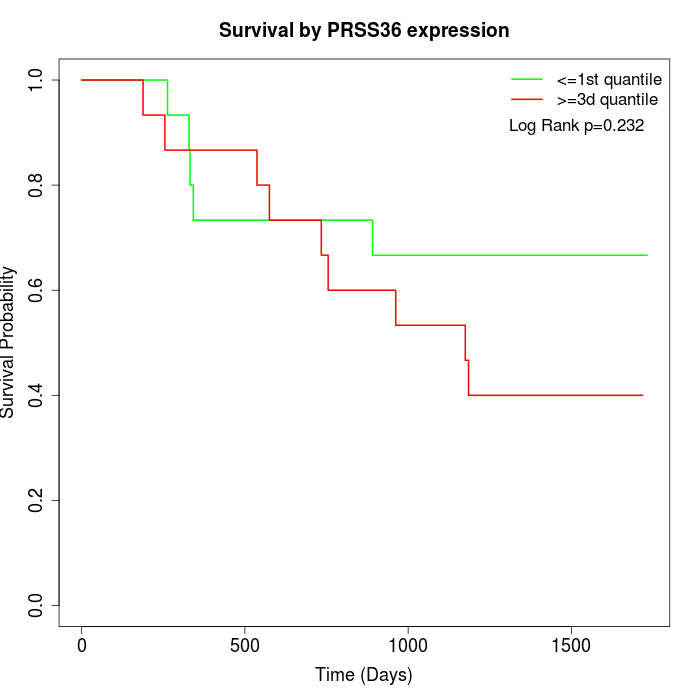
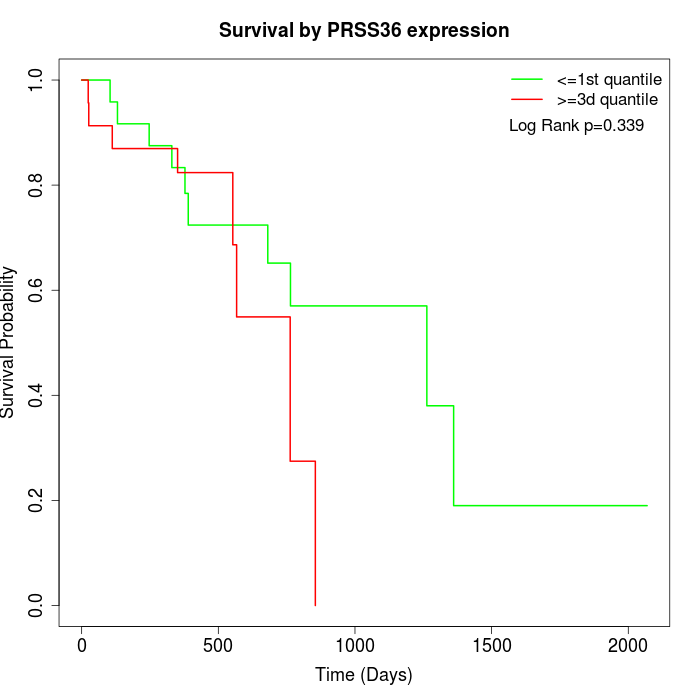
Survival by PRSS36 expression:
Note: Click image to view full size file.
Copy number change of PRSS36:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRSS36 | 146547 | 4 | 5 | 21 | |
| GSE20123 | PRSS36 | 146547 | 4 | 4 | 22 | |
| GSE43470 | PRSS36 | 146547 | 3 | 3 | 37 | |
| GSE46452 | PRSS36 | 146547 | 38 | 1 | 20 | |
| GSE47630 | PRSS36 | 146547 | 11 | 7 | 22 | |
| GSE54993 | PRSS36 | 146547 | 2 | 5 | 63 | |
| GSE54994 | PRSS36 | 146547 | 5 | 9 | 39 | |
| GSE60625 | PRSS36 | 146547 | 4 | 0 | 7 | |
| GSE74703 | PRSS36 | 146547 | 3 | 2 | 31 | |
| GSE74704 | PRSS36 | 146547 | 3 | 2 | 15 | |
| TCGA | PRSS36 | 146547 | 20 | 10 | 66 |
Total number of gains: 97; Total number of losses: 48; Total Number of normals: 343.
Somatic mutations of PRSS36:
Generating mutation plots.
Highly correlated genes for PRSS36:
Showing top 20/245 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRSS36 | LRRD1 | 0.868573 | 3 | 0 | 3 |
| PRSS36 | KRTAP19-2 | 0.85646 | 3 | 0 | 3 |
| PRSS36 | SPATA21 | 0.813125 | 3 | 0 | 3 |
| PRSS36 | RFTN2 | 0.809935 | 3 | 0 | 3 |
| PRSS36 | FOXP4 | 0.809197 | 3 | 0 | 3 |
| PRSS36 | KRTAP21-2 | 0.801538 | 3 | 0 | 3 |
| PRSS36 | APOB | 0.77371 | 3 | 0 | 3 |
| PRSS36 | USP40 | 0.754411 | 3 | 0 | 3 |
| PRSS36 | MEX3B | 0.753268 | 3 | 0 | 3 |
| PRSS36 | ZNF878 | 0.739318 | 3 | 0 | 3 |
| PRSS36 | RAB4B | 0.738955 | 3 | 0 | 3 |
| PRSS36 | ZNF546 | 0.738809 | 3 | 0 | 3 |
| PRSS36 | SFTPA2 | 0.738526 | 3 | 0 | 3 |
| PRSS36 | FAM167B | 0.735692 | 5 | 0 | 5 |
| PRSS36 | SERPINA1 | 0.733237 | 3 | 0 | 3 |
| PRSS36 | C5AR1 | 0.73234 | 3 | 0 | 3 |
| PRSS36 | VCP | 0.731844 | 3 | 0 | 3 |
| PRSS36 | KIR3DL2 | 0.72837 | 3 | 0 | 3 |
| PRSS36 | MED22 | 0.727874 | 3 | 0 | 3 |
| PRSS36 | ERF | 0.724507 | 3 | 0 | 3 |
For details and further investigation, click here