| Full name: MIR7-3 host gene | Alias Symbol: PGSF1|PGSF1a|PGSF1b|Huh7|uc002mbe.2 | ||
| Type: non-coding RNA | Cytoband: 19p13.3 | ||
| Entrez ID: 284424 | HGNC ID: HGNC:30049 | Ensembl Gene: ENSG00000176840 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of MIR7-3HG:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MIR7-3HG | 284424 | 223913_s_at | -0.0402 | 0.8816 | |
| GSE26886 | MIR7-3HG | 284424 | 223913_s_at | 0.0479 | 0.5969 | |
| GSE45670 | MIR7-3HG | 284424 | 223913_s_at | 0.0698 | 0.4412 | |
| GSE53622 | MIR7-3HG | 284424 | 35888 | 0.4832 | 0.0000 | |
| GSE53624 | MIR7-3HG | 284424 | 35888 | 1.1478 | 0.0000 | |
| GSE63941 | MIR7-3HG | 284424 | 223973_at | 0.0957 | 0.5121 | |
| GSE77861 | MIR7-3HG | 284424 | 223973_at | -0.1726 | 0.1352 |
Upregulated datasets: 1; Downregulated datasets: 0.
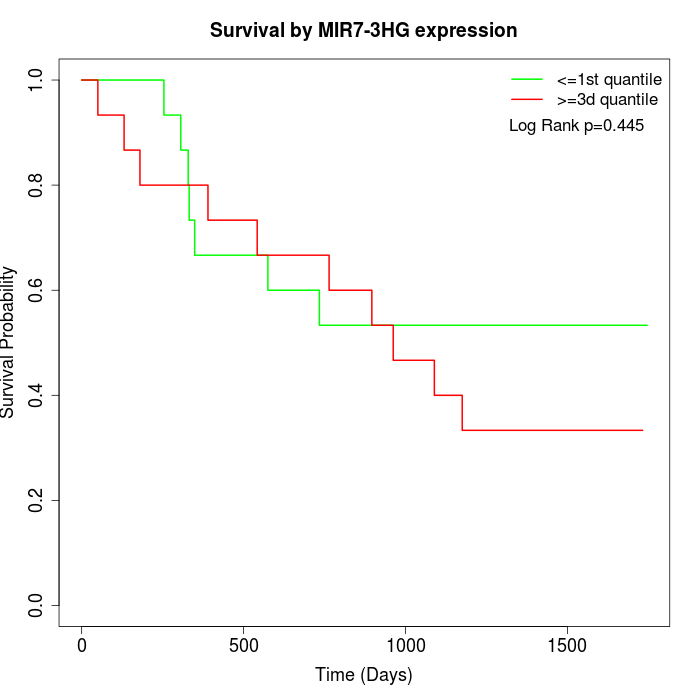
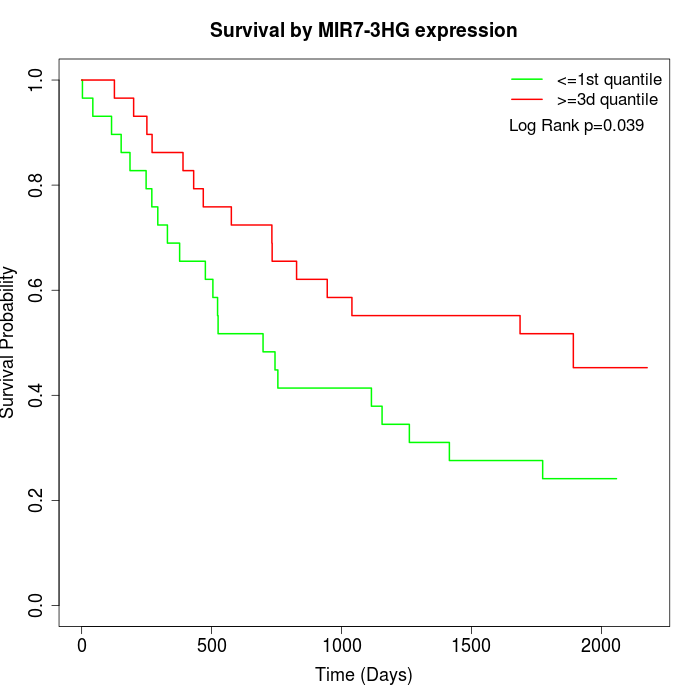
Survival by MIR7-3HG expression:
Note: Click image to view full size file.
Copy number change of MIR7-3HG:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MIR7-3HG | 284424 | 5 | 3 | 22 | |
| GSE20123 | MIR7-3HG | 284424 | 4 | 2 | 24 | |
| GSE43470 | MIR7-3HG | 284424 | 1 | 8 | 34 | |
| GSE46452 | MIR7-3HG | 284424 | 47 | 1 | 11 | |
| GSE47630 | MIR7-3HG | 284424 | 5 | 7 | 28 | |
| GSE54993 | MIR7-3HG | 284424 | 16 | 3 | 51 | |
| GSE54994 | MIR7-3HG | 284424 | 6 | 16 | 31 | |
| GSE60625 | MIR7-3HG | 284424 | 9 | 0 | 2 | |
| GSE74703 | MIR7-3HG | 284424 | 1 | 5 | 30 | |
| GSE74704 | MIR7-3HG | 284424 | 1 | 2 | 17 |
Total number of gains: 95; Total number of losses: 47; Total Number of normals: 250.
Somatic mutations of MIR7-3HG:
Generating mutation plots.
Highly correlated genes for MIR7-3HG:
Showing top 20/65 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MIR7-3HG | CALCA | 0.700859 | 3 | 0 | 3 |
| MIR7-3HG | OVCH1-AS1 | 0.685975 | 3 | 0 | 3 |
| MIR7-3HG | LINC00868 | 0.678853 | 3 | 0 | 3 |
| MIR7-3HG | TSPO2 | 0.674896 | 3 | 0 | 3 |
| MIR7-3HG | FAM71A | 0.668625 | 3 | 0 | 3 |
| MIR7-3HG | LINC01015 | 0.65985 | 3 | 0 | 3 |
| MIR7-3HG | CKM | 0.651924 | 3 | 0 | 3 |
| MIR7-3HG | ZNF469 | 0.647503 | 3 | 0 | 3 |
| MIR7-3HG | WWC2-AS2 | 0.641941 | 3 | 0 | 3 |
| MIR7-3HG | NANOG | 0.617468 | 3 | 0 | 3 |
| MIR7-3HG | FNDC1 | 0.615009 | 3 | 0 | 3 |
| MIR7-3HG | AMHR2 | 0.613773 | 3 | 0 | 3 |
| MIR7-3HG | MYLK2 | 0.607676 | 4 | 0 | 4 |
| MIR7-3HG | TSSK6 | 0.605532 | 4 | 0 | 4 |
| MIR7-3HG | GPR32 | 0.604516 | 4 | 0 | 3 |
| MIR7-3HG | VWA5B2 | 0.584349 | 4 | 0 | 3 |
| MIR7-3HG | GUCA1A | 0.582565 | 4 | 0 | 3 |
| MIR7-3HG | F11 | 0.578834 | 3 | 0 | 3 |
| MIR7-3HG | ZNRF4 | 0.57798 | 5 | 0 | 3 |
| MIR7-3HG | APOA5 | 0.577232 | 3 | 0 | 3 |
For details and further investigation, click here