| Full name: creatine kinase, M-type | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 19q13.32 | ||
| Entrez ID: 1158 | HGNC ID: HGNC:1994 | Ensembl Gene: ENSG00000104879 | OMIM ID: 123310 |
| Drug and gene relationship at DGIdb | |||
Expression of CKM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CKM | 1158 | 204810_s_at | -0.0079 | 0.9799 | |
| GSE20347 | CKM | 1158 | 204810_s_at | -0.0499 | 0.6884 | |
| GSE23400 | CKM | 1158 | 204810_s_at | -0.1049 | 0.3417 | |
| GSE26886 | CKM | 1158 | 204810_s_at | 0.0527 | 0.7097 | |
| GSE29001 | CKM | 1158 | 204810_s_at | -0.1113 | 0.3708 | |
| GSE38129 | CKM | 1158 | 204810_s_at | -0.3371 | 0.0867 | |
| GSE45670 | CKM | 1158 | 204810_s_at | 0.1175 | 0.2593 | |
| GSE53622 | CKM | 1158 | 56535 | -0.3542 | 0.0250 | |
| GSE53624 | CKM | 1158 | 56535 | -0.2085 | 0.0481 | |
| GSE63941 | CKM | 1158 | 204810_s_at | 0.3884 | 0.3768 | |
| GSE77861 | CKM | 1158 | 204810_s_at | -0.2155 | 0.0560 | |
| GSE97050 | CKM | 1158 | A_23_P50250 | -2.2089 | 0.1421 | |
| TCGA | CKM | 1158 | RNAseq | -3.5059 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
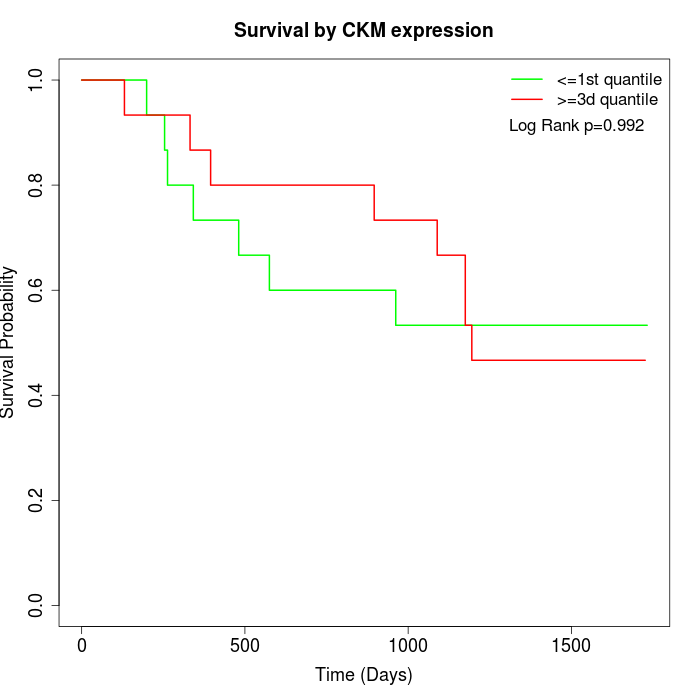
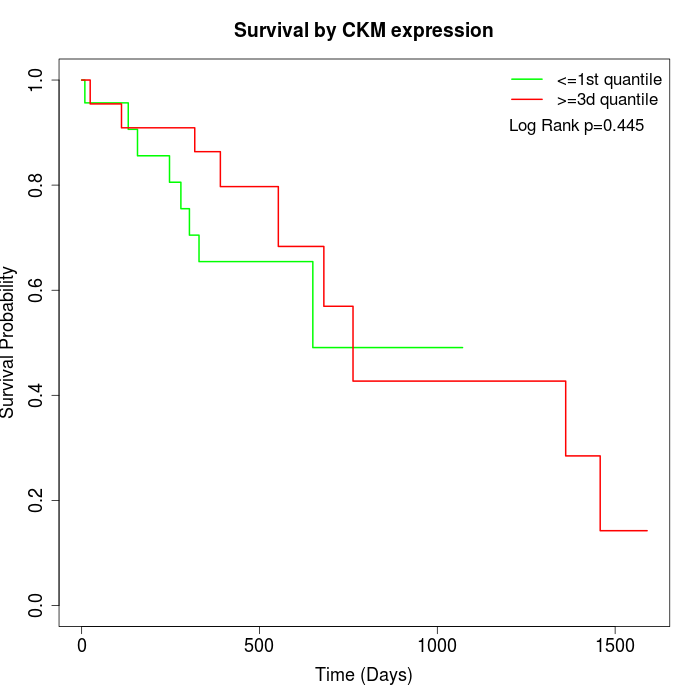
Survival by CKM expression:
Note: Click image to view full size file.
Copy number change of CKM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CKM | 1158 | 4 | 5 | 21 | |
| GSE20123 | CKM | 1158 | 4 | 4 | 22 | |
| GSE43470 | CKM | 1158 | 3 | 11 | 29 | |
| GSE46452 | CKM | 1158 | 45 | 1 | 13 | |
| GSE47630 | CKM | 1158 | 8 | 6 | 26 | |
| GSE54993 | CKM | 1158 | 17 | 4 | 49 | |
| GSE54994 | CKM | 1158 | 6 | 12 | 35 | |
| GSE60625 | CKM | 1158 | 9 | 0 | 2 | |
| GSE74703 | CKM | 1158 | 3 | 7 | 26 | |
| GSE74704 | CKM | 1158 | 4 | 2 | 14 | |
| TCGA | CKM | 1158 | 15 | 16 | 65 |
Total number of gains: 118; Total number of losses: 68; Total Number of normals: 302.
Somatic mutations of CKM:
Generating mutation plots.
Highly correlated genes for CKM:
Showing top 20/335 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CKM | TCAP | 0.812305 | 4 | 0 | 4 |
| CKM | SIX6 | 0.725276 | 3 | 0 | 3 |
| CKM | TSGA13 | 0.719562 | 3 | 0 | 3 |
| CKM | MYL1 | 0.713163 | 4 | 0 | 3 |
| CKM | CSRP3 | 0.702549 | 5 | 0 | 5 |
| CKM | REG1A | 0.701945 | 3 | 0 | 3 |
| CKM | TTN | 0.701795 | 5 | 0 | 4 |
| CKM | UBQLN3 | 0.692176 | 3 | 0 | 3 |
| CKM | IFNA10 | 0.689067 | 3 | 0 | 3 |
| CKM | AFF3 | 0.688942 | 3 | 0 | 3 |
| CKM | CASC2 | 0.687504 | 3 | 0 | 3 |
| CKM | DLGAP2 | 0.687388 | 3 | 0 | 3 |
| CKM | ASIC2 | 0.684166 | 3 | 0 | 3 |
| CKM | ITIH1 | 0.681825 | 3 | 0 | 3 |
| CKM | TFDP3 | 0.677389 | 3 | 0 | 3 |
| CKM | CCDC60 | 0.669696 | 3 | 0 | 3 |
| CKM | GH2 | 0.667964 | 4 | 0 | 4 |
| CKM | LINC00868 | 0.667365 | 3 | 0 | 3 |
| CKM | CSDC2 | 0.660197 | 4 | 0 | 4 |
| CKM | GJD2 | 0.660083 | 4 | 0 | 4 |
For details and further investigation, click here