| Full name: myosin light chain 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 2q34 | ||
| Entrez ID: 4632 | HGNC ID: HGNC:7582 | Ensembl Gene: ENSG00000168530 | OMIM ID: 160780 |
| Drug and gene relationship at DGIdb | |||
Expression of MYL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MYL1 | 4632 | 209888_s_at | -0.0641 | 0.7822 | |
| GSE20347 | MYL1 | 4632 | 209888_s_at | 0.0209 | 0.8167 | |
| GSE23400 | MYL1 | 4632 | 209888_s_at | 0.1091 | 0.2210 | |
| GSE26886 | MYL1 | 4632 | 209888_s_at | -0.0173 | 0.9142 | |
| GSE29001 | MYL1 | 4632 | 209888_s_at | -0.0542 | 0.7405 | |
| GSE38129 | MYL1 | 4632 | 209888_s_at | -0.1030 | 0.2451 | |
| GSE45670 | MYL1 | 4632 | 209888_s_at | 0.0019 | 0.9886 | |
| GSE53622 | MYL1 | 4632 | 79685 | 0.0203 | 0.9227 | |
| GSE53624 | MYL1 | 4632 | 79685 | 0.0073 | 0.9622 | |
| GSE63941 | MYL1 | 4632 | 209888_s_at | 0.0353 | 0.8035 | |
| GSE77861 | MYL1 | 4632 | 209888_s_at | -0.0321 | 0.8064 | |
| GSE97050 | MYL1 | 4632 | A_24_P191326 | -1.6446 | 0.2122 |
Upregulated datasets: 0; Downregulated datasets: 0.
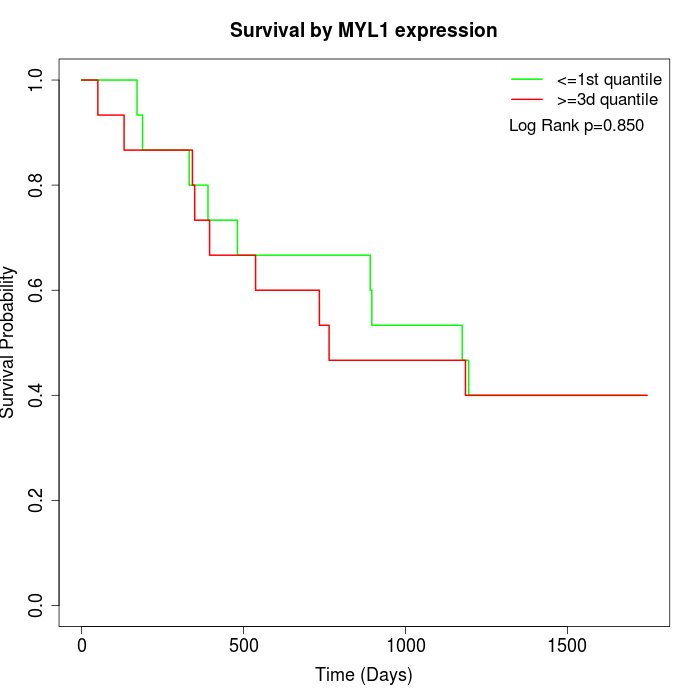
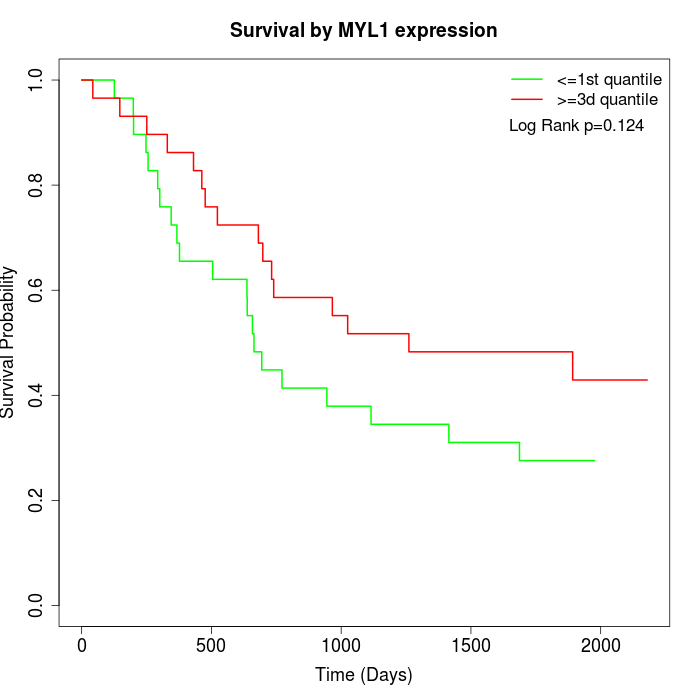
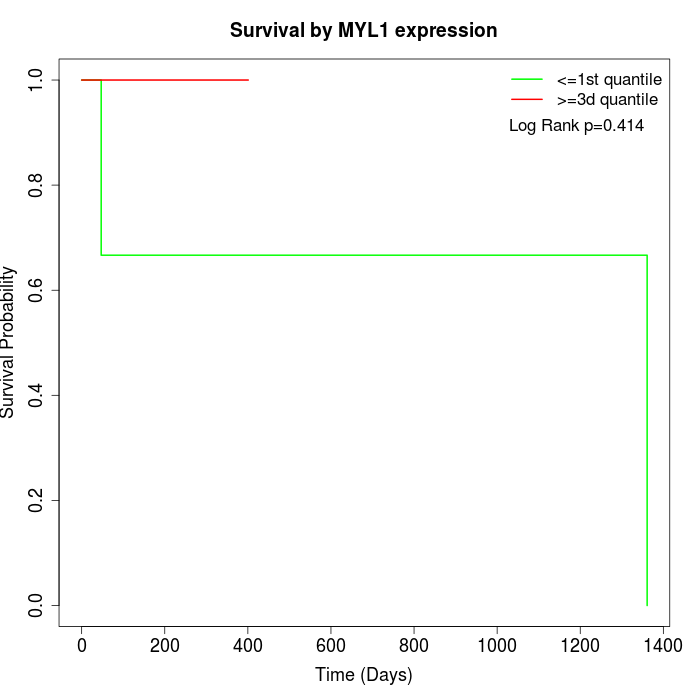
Survival by MYL1 expression:
Note: Click image to view full size file.
Copy number change of MYL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MYL1 | 4632 | 2 | 12 | 16 | |
| GSE20123 | MYL1 | 4632 | 2 | 12 | 16 | |
| GSE43470 | MYL1 | 4632 | 1 | 6 | 36 | |
| GSE46452 | MYL1 | 4632 | 0 | 5 | 54 | |
| GSE47630 | MYL1 | 4632 | 4 | 5 | 31 | |
| GSE54993 | MYL1 | 4632 | 0 | 3 | 67 | |
| GSE54994 | MYL1 | 4632 | 9 | 9 | 35 | |
| GSE60625 | MYL1 | 4632 | 0 | 3 | 8 | |
| GSE74703 | MYL1 | 4632 | 1 | 5 | 30 | |
| GSE74704 | MYL1 | 4632 | 2 | 6 | 12 | |
| TCGA | MYL1 | 4632 | 12 | 25 | 59 |
Total number of gains: 33; Total number of losses: 91; Total Number of normals: 364.
Somatic mutations of MYL1:
Generating mutation plots.
Highly correlated genes for MYL1:
Showing top 20/653 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MYL1 | FOXN3-AS2 | 0.754963 | 4 | 0 | 4 |
| MYL1 | CACNG2 | 0.754634 | 3 | 0 | 3 |
| MYL1 | CAMKV | 0.747416 | 3 | 0 | 3 |
| MYL1 | PRORY | 0.726181 | 4 | 0 | 4 |
| MYL1 | NUDT7 | 0.724732 | 3 | 0 | 3 |
| MYL1 | FXYD7 | 0.72275 | 4 | 0 | 4 |
| MYL1 | MYL2 | 0.721431 | 4 | 0 | 4 |
| MYL1 | STAT2 | 0.716646 | 3 | 0 | 3 |
| MYL1 | IL25 | 0.713979 | 3 | 0 | 3 |
| MYL1 | CKM | 0.713163 | 4 | 0 | 3 |
| MYL1 | CNGA3 | 0.712819 | 3 | 0 | 3 |
| MYL1 | CCDC87 | 0.712537 | 3 | 0 | 3 |
| MYL1 | HEYL | 0.706123 | 3 | 0 | 3 |
| MYL1 | KIR2DS5 | 0.704286 | 4 | 0 | 3 |
| MYL1 | IL18BP | 0.704003 | 3 | 0 | 3 |
| MYL1 | NTNG1 | 0.703555 | 3 | 0 | 3 |
| MYL1 | NKX2-1 | 0.699084 | 3 | 0 | 3 |
| MYL1 | BCAN | 0.697709 | 5 | 0 | 5 |
| MYL1 | MTAP | 0.695634 | 3 | 0 | 3 |
| MYL1 | TRIM10 | 0.694429 | 4 | 0 | 4 |
For details and further investigation, click here