| Full name: NK2 homeobox 1 | Alias Symbol: TTF-1|TTF1 | ||
| Type: protein-coding gene | Cytoband: 14q13.3 | ||
| Entrez ID: 7080 | HGNC ID: HGNC:11825 | Ensembl Gene: ENSG00000136352 | OMIM ID: 600635 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of NKX2-1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NKX2-1 | 7080 | 210673_x_at | -0.0505 | 0.8776 | |
| GSE20347 | NKX2-1 | 7080 | 210673_x_at | -0.1599 | 0.0217 | |
| GSE23400 | NKX2-1 | 7080 | 210673_x_at | -0.2404 | 0.0000 | |
| GSE26886 | NKX2-1 | 7080 | 210673_x_at | 0.0206 | 0.9093 | |
| GSE29001 | NKX2-1 | 7080 | 211024_s_at | 0.0464 | 0.7545 | |
| GSE38129 | NKX2-1 | 7080 | 210673_x_at | -0.0146 | 0.9035 | |
| GSE45670 | NKX2-1 | 7080 | 210673_x_at | 0.0318 | 0.7865 | |
| GSE53622 | NKX2-1 | 7080 | 14539 | 0.9888 | 0.0000 | |
| GSE53624 | NKX2-1 | 7080 | 14539 | 1.5472 | 0.0000 | |
| GSE63941 | NKX2-1 | 7080 | 210673_x_at | 0.3822 | 0.0835 | |
| GSE77861 | NKX2-1 | 7080 | 210673_x_at | -0.1334 | 0.4693 | |
| TCGA | NKX2-1 | 7080 | RNAseq | 3.6389 | 0.0010 |
Upregulated datasets: 2; Downregulated datasets: 0.
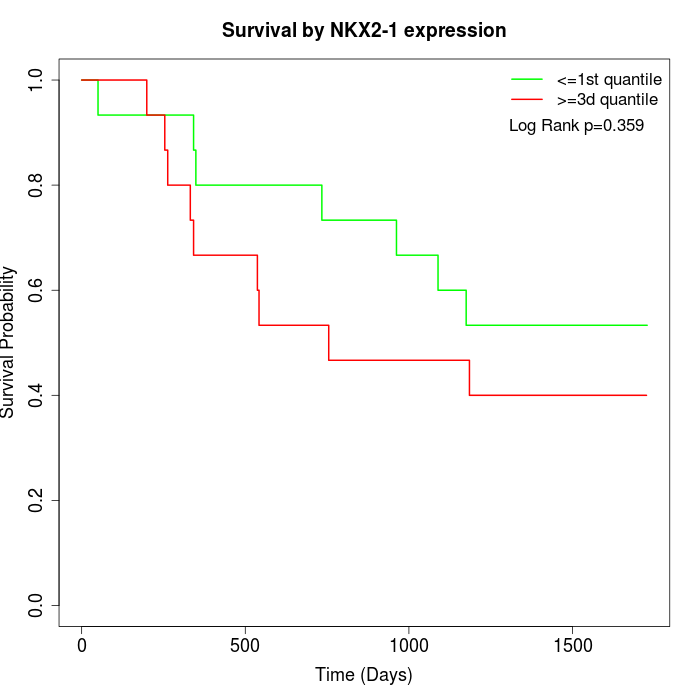
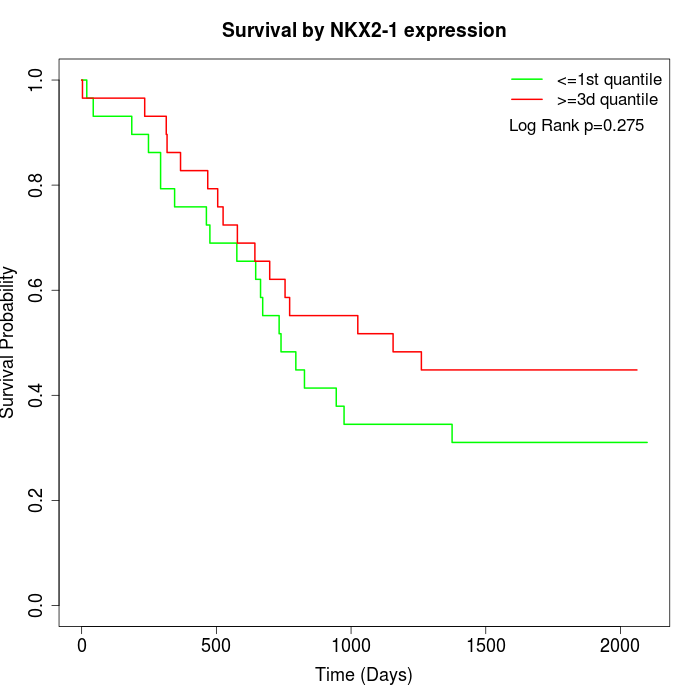
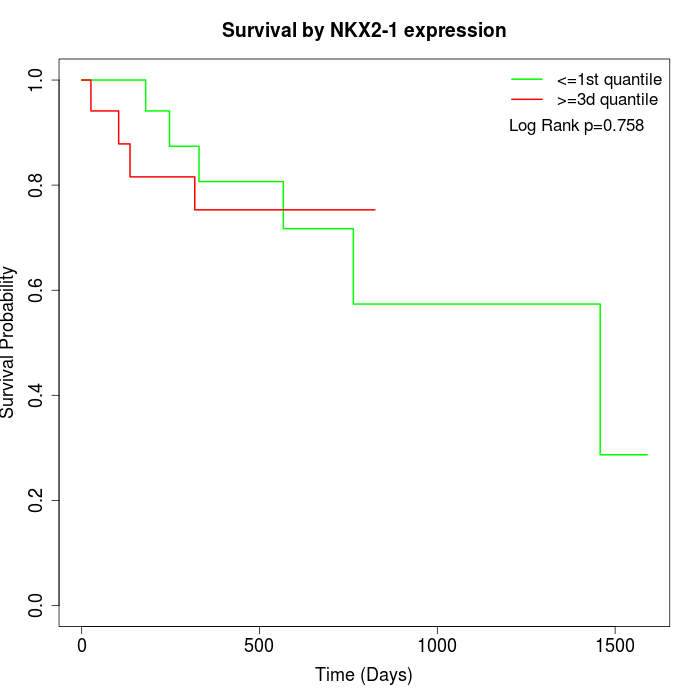
Survival by NKX2-1 expression:
Note: Click image to view full size file.
Copy number change of NKX2-1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NKX2-1 | 7080 | 11 | 4 | 15 | |
| GSE20123 | NKX2-1 | 7080 | 12 | 4 | 14 | |
| GSE43470 | NKX2-1 | 7080 | 8 | 2 | 33 | |
| GSE46452 | NKX2-1 | 7080 | 18 | 2 | 39 | |
| GSE47630 | NKX2-1 | 7080 | 11 | 10 | 19 | |
| GSE54993 | NKX2-1 | 7080 | 3 | 11 | 56 | |
| GSE54994 | NKX2-1 | 7080 | 19 | 5 | 29 | |
| GSE60625 | NKX2-1 | 7080 | 0 | 2 | 9 | |
| GSE74703 | NKX2-1 | 7080 | 7 | 2 | 27 | |
| GSE74704 | NKX2-1 | 7080 | 6 | 3 | 11 |
Total number of gains: 95; Total number of losses: 45; Total Number of normals: 252.
Somatic mutations of NKX2-1:
Generating mutation plots.
Highly correlated genes for NKX2-1:
Showing top 20/527 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NKX2-1 | LOXHD1 | 0.746166 | 3 | 0 | 3 |
| NKX2-1 | MYL1 | 0.699084 | 3 | 0 | 3 |
| NKX2-1 | SNX8 | 0.681982 | 4 | 0 | 3 |
| NKX2-1 | DNAJB7 | 0.680568 | 3 | 0 | 3 |
| NKX2-1 | ADAM15 | 0.679417 | 3 | 0 | 3 |
| NKX2-1 | SMYD1 | 0.677558 | 3 | 0 | 3 |
| NKX2-1 | MYT1 | 0.671473 | 4 | 0 | 4 |
| NKX2-1 | NGB | 0.671207 | 5 | 0 | 5 |
| NKX2-1 | FAM131C | 0.658019 | 3 | 0 | 3 |
| NKX2-1 | BIRC7 | 0.654308 | 6 | 0 | 5 |
| NKX2-1 | ANKRD53 | 0.653249 | 3 | 0 | 3 |
| NKX2-1 | KCNQ4 | 0.651917 | 6 | 0 | 6 |
| NKX2-1 | SARDH | 0.64903 | 5 | 0 | 5 |
| NKX2-1 | SLC6A11 | 0.648407 | 5 | 0 | 5 |
| NKX2-1 | PRKAR1B | 0.646556 | 3 | 0 | 3 |
| NKX2-1 | CELF5 | 0.639557 | 3 | 0 | 3 |
| NKX2-1 | PRR29 | 0.629532 | 4 | 0 | 3 |
| NKX2-1 | OVGP1 | 0.628051 | 3 | 0 | 3 |
| NKX2-1 | RFX2 | 0.625737 | 3 | 0 | 3 |
| NKX2-1 | DRP2 | 0.625343 | 4 | 0 | 4 |
For details and further investigation, click here