| Full name: methylthioadenosine phosphorylase | Alias Symbol: MSAP|c86fus | ||
| Type: protein-coding gene | Cytoband: 9p21.3 | ||
| Entrez ID: 4507 | HGNC ID: HGNC:7413 | Ensembl Gene: ENSG00000099810 | OMIM ID: 156540 |
| Drug and gene relationship at DGIdb | |||
Expression of MTAP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MTAP | 4507 | 231984_at | -0.7743 | 0.2157 | |
| GSE20347 | MTAP | 4507 | 211364_at | 0.0582 | 0.6678 | |
| GSE23400 | MTAP | 4507 | 211364_at | -0.2082 | 0.0000 | |
| GSE26886 | MTAP | 4507 | 231984_at | 0.8353 | 0.0033 | |
| GSE29001 | MTAP | 4507 | 211364_at | -0.1857 | 0.3153 | |
| GSE38129 | MTAP | 4507 | 211364_at | -0.0931 | 0.4494 | |
| GSE45670 | MTAP | 4507 | 231984_at | -0.2686 | 0.6471 | |
| GSE53622 | MTAP | 4507 | 53144 | -0.2649 | 0.1845 | |
| GSE53624 | MTAP | 4507 | 157100 | -0.4549 | 0.0045 | |
| GSE63941 | MTAP | 4507 | 231984_at | -0.7893 | 0.6154 | |
| GSE77861 | MTAP | 4507 | 216685_s_at | -0.0901 | 0.8017 | |
| GSE97050 | MTAP | 4507 | A_24_P51118 | 0.0738 | 0.9323 | |
| SRP007169 | MTAP | 4507 | RNAseq | 1.1687 | 0.0457 | |
| SRP008496 | MTAP | 4507 | RNAseq | 1.3806 | 0.0018 | |
| SRP064894 | MTAP | 4507 | RNAseq | 0.6297 | 0.0239 | |
| SRP133303 | MTAP | 4507 | RNAseq | 0.1964 | 0.6352 | |
| SRP159526 | MTAP | 4507 | RNAseq | -0.3031 | 0.6449 | |
| SRP193095 | MTAP | 4507 | RNAseq | 0.5626 | 0.0650 | |
| SRP219564 | MTAP | 4507 | RNAseq | -0.4413 | 0.3752 | |
| TCGA | MTAP | 4507 | RNAseq | -0.0248 | 0.8839 |
Upregulated datasets: 2; Downregulated datasets: 0.
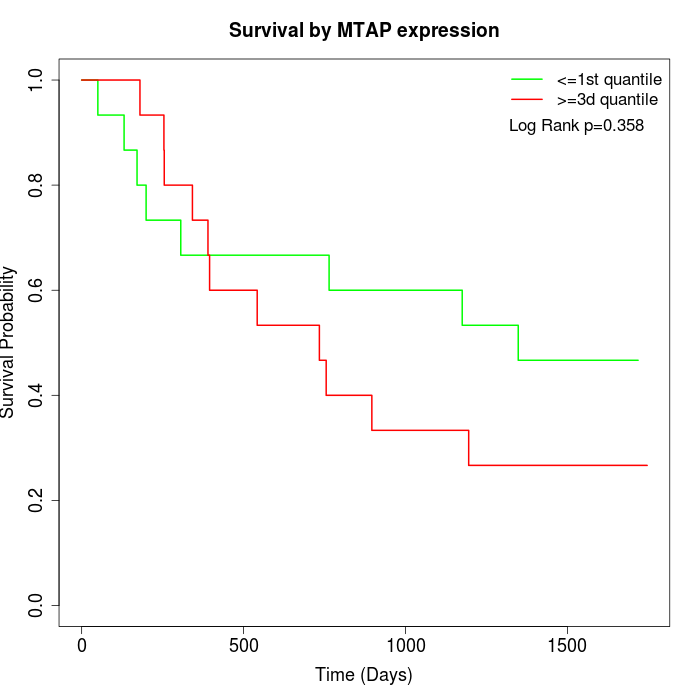
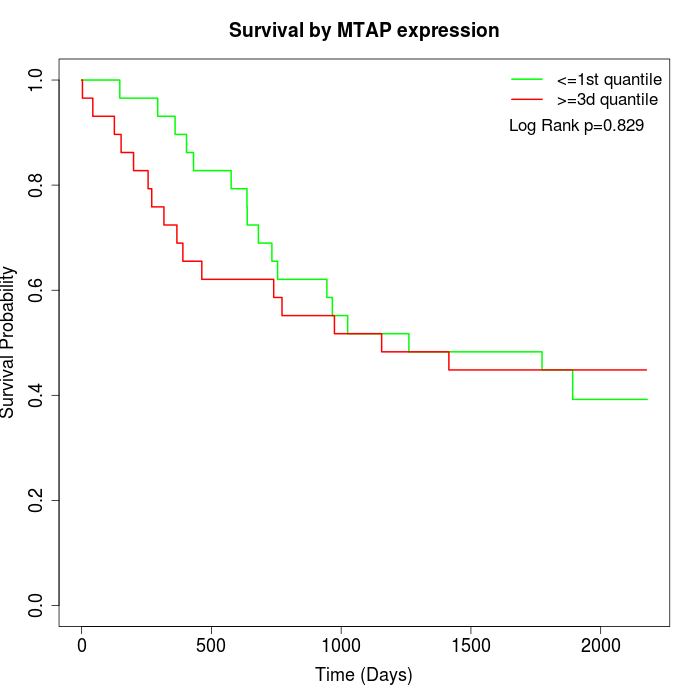
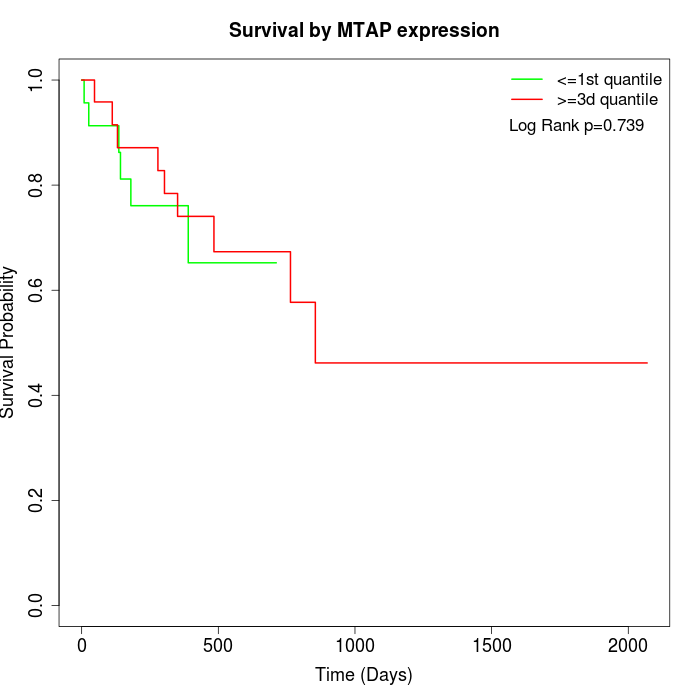
Survival by MTAP expression:
Note: Click image to view full size file.
Copy number change of MTAP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MTAP | 4507 | 1 | 22 | 7 | |
| GSE20123 | MTAP | 4507 | 1 | 22 | 7 | |
| GSE43470 | MTAP | 4507 | 3 | 14 | 26 | |
| GSE46452 | MTAP | 4507 | 1 | 28 | 30 | |
| GSE47630 | MTAP | 4507 | 3 | 27 | 10 | |
| GSE54993 | MTAP | 4507 | 9 | 1 | 60 | |
| GSE54994 | MTAP | 4507 | 6 | 25 | 22 | |
| GSE60625 | MTAP | 4507 | 3 | 0 | 8 | |
| GSE74703 | MTAP | 4507 | 3 | 11 | 22 | |
| GSE74704 | MTAP | 4507 | 0 | 15 | 5 | |
| TCGA | MTAP | 4507 | 5 | 69 | 22 |
Total number of gains: 35; Total number of losses: 234; Total Number of normals: 219.
Somatic mutations of MTAP:
Generating mutation plots.
Highly correlated genes for MTAP:
Showing top 20/850 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MTAP | CLC | 0.811113 | 3 | 0 | 3 |
| MTAP | ITGB3 | 0.791512 | 5 | 0 | 5 |
| MTAP | CPA2 | 0.785181 | 3 | 0 | 3 |
| MTAP | PHLDB1 | 0.763484 | 5 | 0 | 5 |
| MTAP | E2F4 | 0.763077 | 5 | 0 | 5 |
| MTAP | GPA33 | 0.760774 | 5 | 0 | 5 |
| MTAP | CER1 | 0.758505 | 5 | 0 | 5 |
| MTAP | ARMCX4 | 0.758341 | 3 | 0 | 3 |
| MTAP | CA7 | 0.75602 | 4 | 0 | 4 |
| MTAP | SST | 0.755826 | 3 | 0 | 3 |
| MTAP | IFNA17 | 0.755751 | 4 | 0 | 4 |
| MTAP | MEFV | 0.755131 | 3 | 0 | 3 |
| MTAP | IFNA14 | 0.747893 | 4 | 0 | 4 |
| MTAP | TP53I11 | 0.744111 | 3 | 0 | 3 |
| MTAP | MED24 | 0.740982 | 5 | 0 | 5 |
| MTAP | PGM5 | 0.740389 | 3 | 0 | 3 |
| MTAP | CETN1 | 0.738421 | 4 | 0 | 4 |
| MTAP | IQCC | 0.738112 | 3 | 0 | 3 |
| MTAP | ADAM22 | 0.737283 | 5 | 0 | 5 |
| MTAP | NUDT16 | 0.737003 | 3 | 0 | 3 |
For details and further investigation, click here