| Full name: sodium/potassium transporting ATPase interacting 3 | Alias Symbol: FLJ39630 | ||
| Type: protein-coding gene | Cytoband: 8q12.3 | ||
| Entrez ID: 286183 | HGNC ID: HGNC:26829 | Ensembl Gene: ENSG00000185942 | OMIM ID: 612872 |
| Drug and gene relationship at DGIdb | |||
Expression of NKAIN3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NKAIN3 | 286183 | 1553241_at | -0.0380 | 0.8968 | |
| GSE26886 | NKAIN3 | 286183 | 1553241_at | 0.0801 | 0.3377 | |
| GSE45670 | NKAIN3 | 286183 | 1553241_at | 0.0419 | 0.6117 | |
| GSE53622 | NKAIN3 | 286183 | 4335 | -0.7709 | 0.0000 | |
| GSE53624 | NKAIN3 | 286183 | 4335 | -0.9157 | 0.0000 | |
| GSE63941 | NKAIN3 | 286183 | 1553241_at | 0.3711 | 0.0138 | |
| GSE77861 | NKAIN3 | 286183 | 1553241_at | -0.0065 | 0.9517 | |
| SRP133303 | NKAIN3 | 286183 | RNAseq | -1.2475 | 0.0327 | |
| TCGA | NKAIN3 | 286183 | RNAseq | -2.0976 | 0.1929 |
Upregulated datasets: 0; Downregulated datasets: 1.
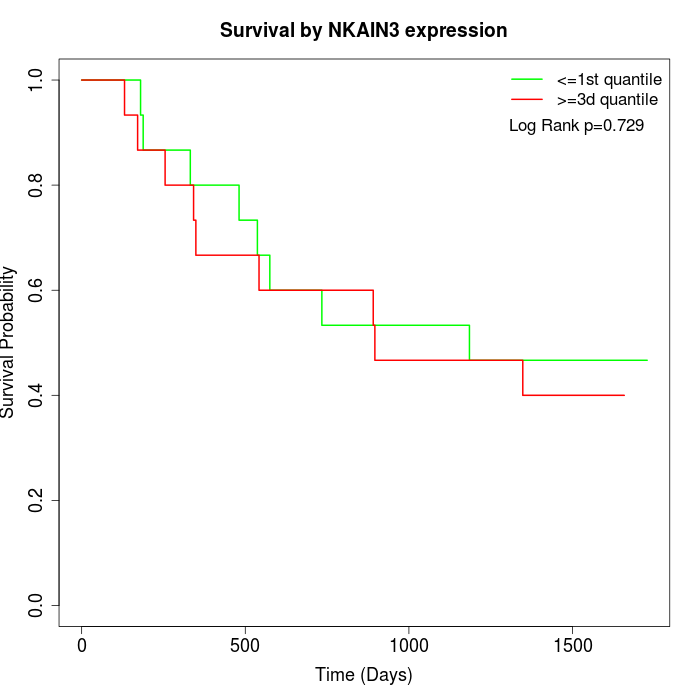
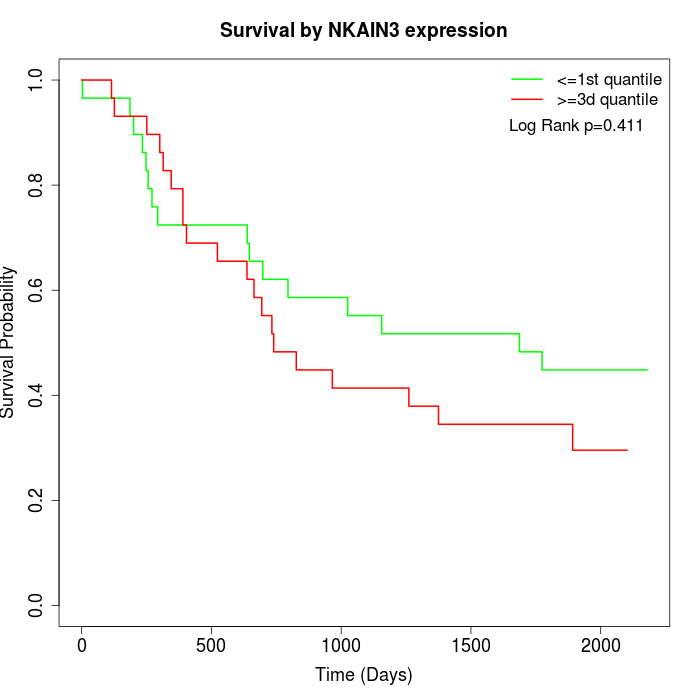
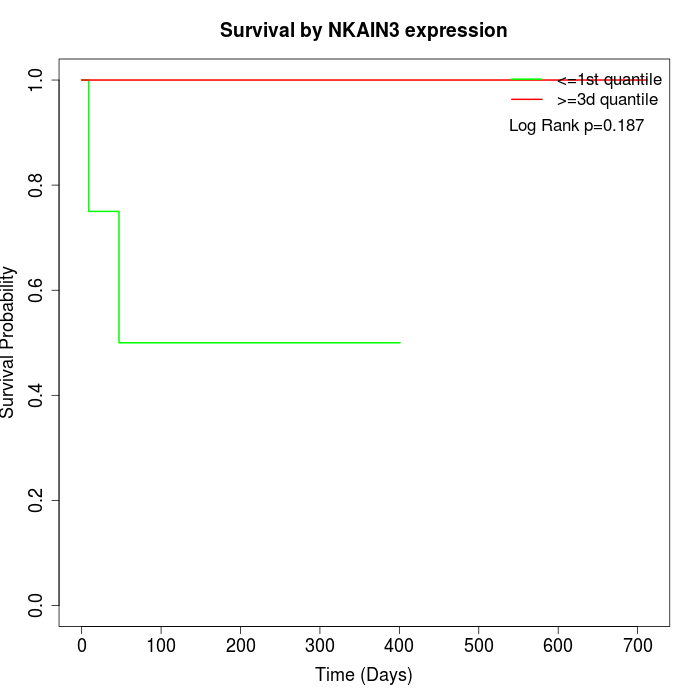
Survival by NKAIN3 expression:
Note: Click image to view full size file.
Copy number change of NKAIN3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NKAIN3 | 286183 | 13 | 1 | 16 | |
| GSE20123 | NKAIN3 | 286183 | 13 | 1 | 16 | |
| GSE43470 | NKAIN3 | 286183 | 15 | 3 | 25 | |
| GSE46452 | NKAIN3 | 286183 | 21 | 1 | 37 | |
| GSE47630 | NKAIN3 | 286183 | 23 | 1 | 16 | |
| GSE54993 | NKAIN3 | 286183 | 1 | 18 | 51 | |
| GSE54994 | NKAIN3 | 286183 | 30 | 0 | 23 | |
| GSE60625 | NKAIN3 | 286183 | 0 | 4 | 7 | |
| GSE74703 | NKAIN3 | 286183 | 13 | 2 | 21 | |
| GSE74704 | NKAIN3 | 286183 | 9 | 0 | 11 | |
| TCGA | NKAIN3 | 286183 | 46 | 6 | 44 |
Total number of gains: 184; Total number of losses: 37; Total Number of normals: 267.
Somatic mutations of NKAIN3:
Generating mutation plots.
Highly correlated genes for NKAIN3:
Showing top 20/33 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NKAIN3 | FAM9C | 0.683352 | 3 | 0 | 3 |
| NKAIN3 | STX18-AS1 | 0.670867 | 3 | 0 | 3 |
| NKAIN3 | SRCIN1 | 0.662166 | 3 | 0 | 3 |
| NKAIN3 | CYLC1 | 0.654529 | 3 | 0 | 3 |
| NKAIN3 | SLC17A4 | 0.61559 | 4 | 0 | 3 |
| NKAIN3 | BAIAP3 | 0.612125 | 3 | 0 | 3 |
| NKAIN3 | SLITRK1 | 0.608489 | 3 | 0 | 3 |
| NKAIN3 | GDF2 | 0.59374 | 3 | 0 | 3 |
| NKAIN3 | GALNT16 | 0.592993 | 4 | 0 | 3 |
| NKAIN3 | RHO | 0.590529 | 4 | 0 | 3 |
| NKAIN3 | LINC01016 | 0.565361 | 6 | 0 | 4 |
| NKAIN3 | DSCAM | 0.563621 | 4 | 0 | 3 |
| NKAIN3 | PRB4 | 0.560508 | 4 | 0 | 3 |
| NKAIN3 | RERGL | 0.5596 | 3 | 0 | 3 |
| NKAIN3 | CAMK2N2 | 0.55675 | 3 | 0 | 3 |
| NKAIN3 | NXPH3 | 0.544634 | 5 | 0 | 3 |
| NKAIN3 | RMST | 0.53031 | 4 | 0 | 3 |
| NKAIN3 | AATK | 0.526026 | 4 | 0 | 3 |
| NKAIN3 | MYBPC3 | 0.525998 | 3 | 0 | 3 |
| NKAIN3 | ASB17 | 0.5251 | 3 | 0 | 3 |
For details and further investigation, click here