| Full name: DS cell adhesion molecule | Alias Symbol: CHD2-42|CHD2-52 | ||
| Type: protein-coding gene | Cytoband: 21q22.2 | ||
| Entrez ID: 1826 | HGNC ID: HGNC:3039 | Ensembl Gene: ENSG00000171587 | OMIM ID: 602523 |
| Drug and gene relationship at DGIdb | |||
Expression of DSCAM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DSCAM | 1826 | 211484_s_at | 0.0999 | 0.7546 | |
| GSE20347 | DSCAM | 1826 | 211484_s_at | 0.1053 | 0.3058 | |
| GSE23400 | DSCAM | 1826 | 211484_s_at | -0.1850 | 0.0000 | |
| GSE26886 | DSCAM | 1826 | 211484_s_at | 0.0403 | 0.7876 | |
| GSE29001 | DSCAM | 1826 | 211484_s_at | -0.1675 | 0.2469 | |
| GSE38129 | DSCAM | 1826 | 211484_s_at | -0.0496 | 0.6825 | |
| GSE45670 | DSCAM | 1826 | 211484_s_at | 0.0125 | 0.9052 | |
| GSE53622 | DSCAM | 1826 | 150808 | 0.0258 | 0.6217 | |
| GSE53624 | DSCAM | 1826 | 150808 | 0.0388 | 0.6679 | |
| GSE63941 | DSCAM | 1826 | 211484_s_at | 0.3073 | 0.0744 | |
| GSE77861 | DSCAM | 1826 | 211484_s_at | 0.0427 | 0.7953 | |
| GSE97050 | DSCAM | 1826 | A_33_P3303305 | -0.0155 | 0.9619 | |
| TCGA | DSCAM | 1826 | RNAseq | 1.4760 | 0.0737 |
Upregulated datasets: 0; Downregulated datasets: 0.
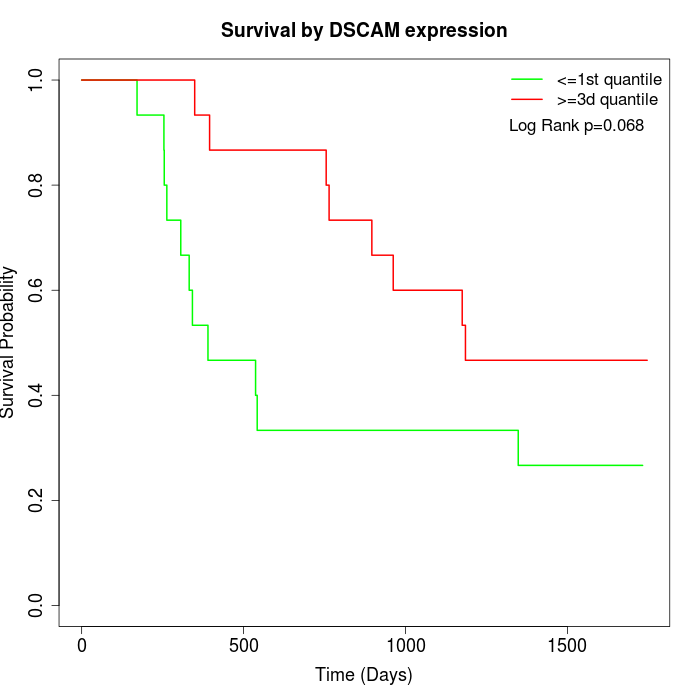
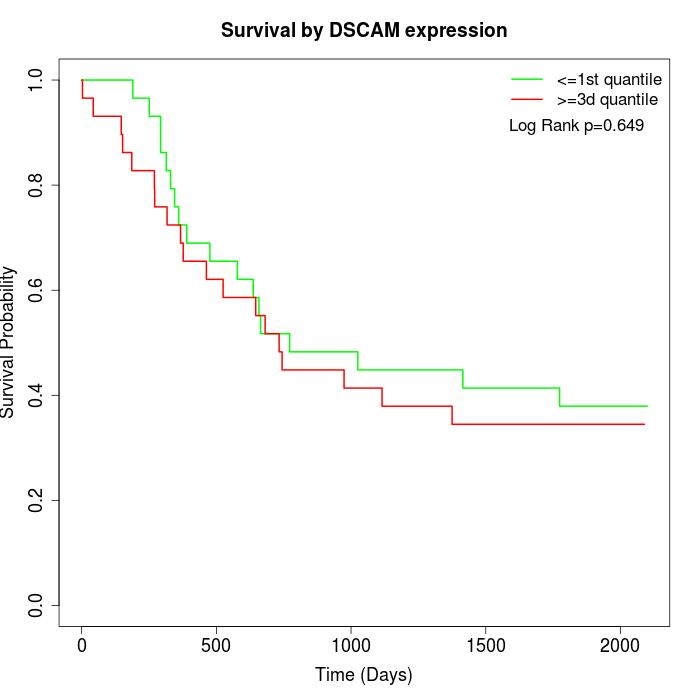
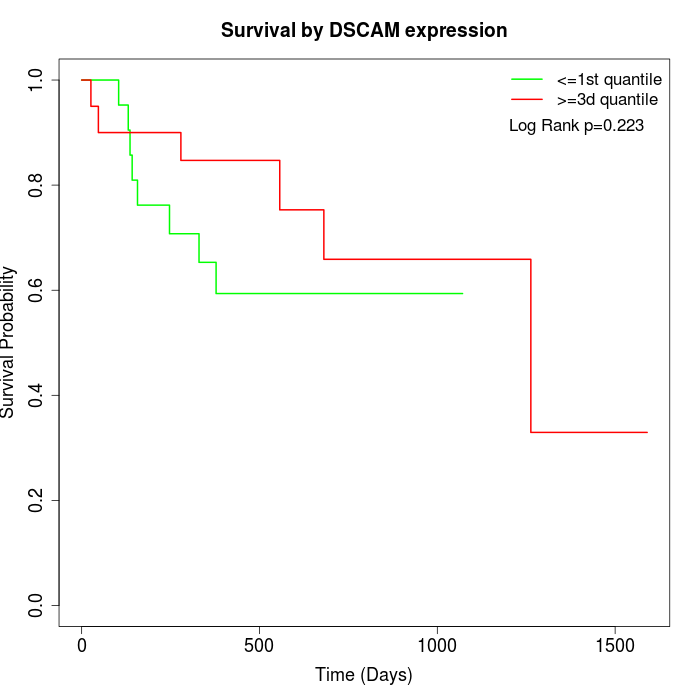
Survival by DSCAM expression:
Note: Click image to view full size file.
Copy number change of DSCAM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DSCAM | 1826 | 2 | 10 | 18 | |
| GSE20123 | DSCAM | 1826 | 2 | 10 | 18 | |
| GSE43470 | DSCAM | 1826 | 1 | 13 | 29 | |
| GSE46452 | DSCAM | 1826 | 1 | 21 | 37 | |
| GSE47630 | DSCAM | 1826 | 6 | 17 | 17 | |
| GSE54993 | DSCAM | 1826 | 8 | 1 | 61 | |
| GSE54994 | DSCAM | 1826 | 3 | 8 | 42 | |
| GSE60625 | DSCAM | 1826 | 0 | 0 | 11 | |
| GSE74703 | DSCAM | 1826 | 1 | 10 | 25 | |
| GSE74704 | DSCAM | 1826 | 1 | 6 | 13 | |
| TCGA | DSCAM | 1826 | 7 | 43 | 46 |
Total number of gains: 32; Total number of losses: 139; Total Number of normals: 317.
Somatic mutations of DSCAM:
Generating mutation plots.
Highly correlated genes for DSCAM:
Showing top 20/1115 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DSCAM | SYCE1L | 0.738248 | 3 | 0 | 3 |
| DSCAM | BEST3 | 0.728385 | 3 | 0 | 3 |
| DSCAM | TXLNB | 0.725468 | 3 | 0 | 3 |
| DSCAM | NKAIN4 | 0.705787 | 3 | 0 | 3 |
| DSCAM | GFRA4 | 0.699614 | 8 | 0 | 7 |
| DSCAM | ZSCAN10 | 0.694601 | 3 | 0 | 3 |
| DSCAM | RUNX2 | 0.689505 | 4 | 0 | 4 |
| DSCAM | TECTA | 0.688964 | 4 | 0 | 4 |
| DSCAM | TMEM235 | 0.683539 | 3 | 0 | 3 |
| DSCAM | CHRNA2 | 0.682827 | 8 | 0 | 8 |
| DSCAM | RASL10A | 0.681001 | 6 | 0 | 6 |
| DSCAM | SLC10A1 | 0.680957 | 7 | 0 | 6 |
| DSCAM | SIRPB2 | 0.680309 | 3 | 0 | 3 |
| DSCAM | IZUMO2 | 0.677462 | 3 | 0 | 3 |
| DSCAM | CACNG4 | 0.676377 | 10 | 0 | 7 |
| DSCAM | OPTC | 0.674324 | 4 | 0 | 4 |
| DSCAM | RHO | 0.673763 | 9 | 0 | 8 |
| DSCAM | CALCA | 0.673063 | 9 | 0 | 8 |
| DSCAM | SLC22A12 | 0.672518 | 3 | 0 | 3 |
| DSCAM | PRB4 | 0.672163 | 7 | 0 | 7 |
For details and further investigation, click here