| Full name: proline rich protein BstNI subfamily 4 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12p13.2 | ||
| Entrez ID: 5545 | HGNC ID: HGNC:9340 | Ensembl Gene: ENSG00000230657 | OMIM ID: 180990 |
| Drug and gene relationship at DGIdb | |||
Expression of PRB4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRB4 | 5545 | 216881_x_at | -0.1621 | 0.4419 | |
| GSE20347 | PRB4 | 5545 | 216881_x_at | 0.0112 | 0.9215 | |
| GSE23400 | PRB4 | 5545 | 216881_x_at | -0.0815 | 0.0300 | |
| GSE26886 | PRB4 | 5545 | 216881_x_at | 0.2336 | 0.1665 | |
| GSE29001 | PRB4 | 5545 | 216881_x_at | -0.1851 | 0.1709 | |
| GSE38129 | PRB4 | 5545 | 216881_x_at | -0.0704 | 0.5085 | |
| GSE45670 | PRB4 | 5545 | 216881_x_at | -0.0682 | 0.4977 | |
| GSE53622 | PRB4 | 5545 | 46759 | -0.6447 | 0.0122 | |
| GSE53624 | PRB4 | 5545 | 46759 | -1.4987 | 0.0000 | |
| GSE63941 | PRB4 | 5545 | 216881_x_at | 0.7375 | 0.0005 | |
| GSE77861 | PRB4 | 5545 | 216881_x_at | -0.1127 | 0.3182 | |
| GSE97050 | PRB4 | 5545 | A_33_P3216150 | -1.4909 | 0.2706 |
Upregulated datasets: 0; Downregulated datasets: 1.
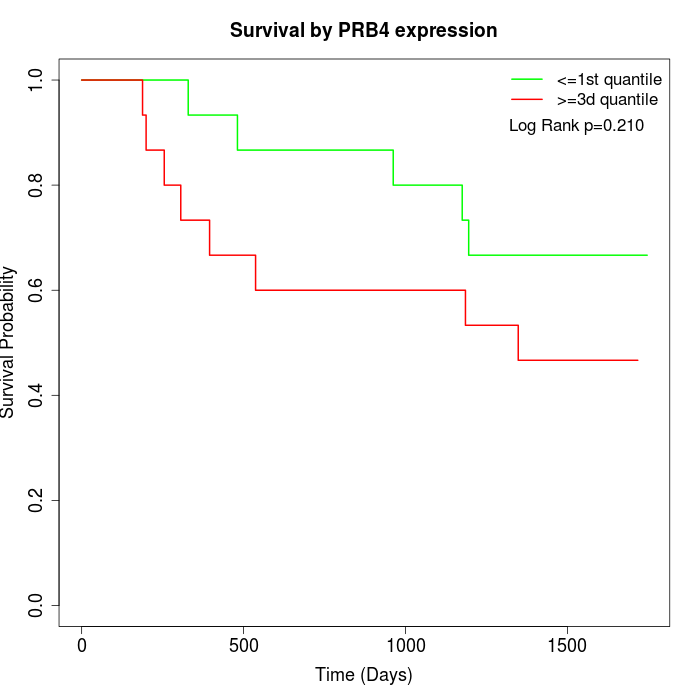
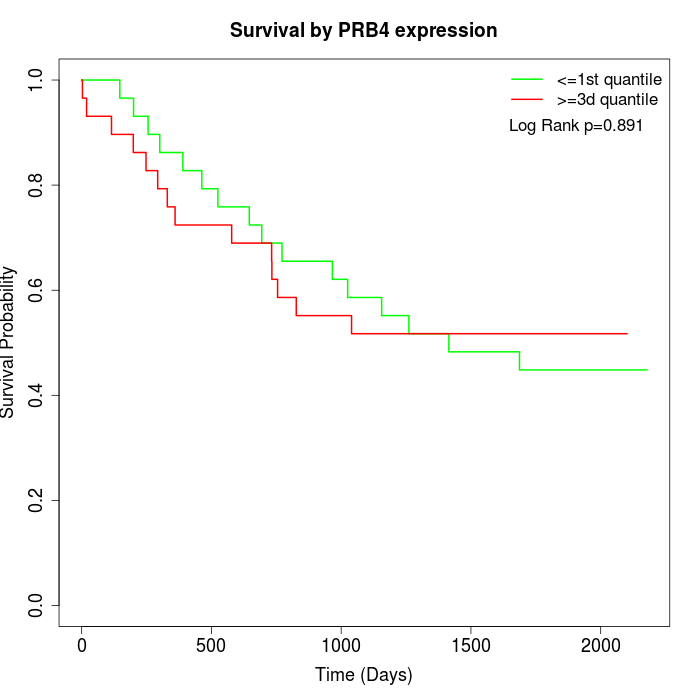
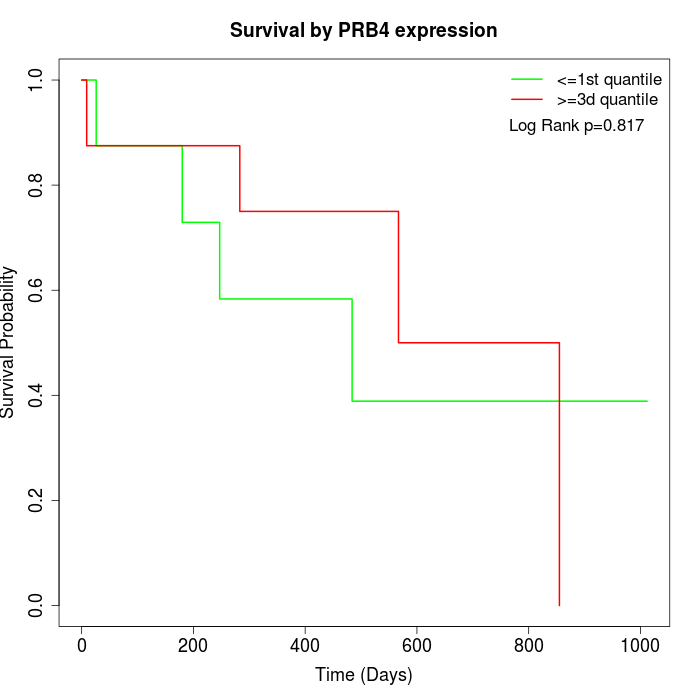
Survival by PRB4 expression:
Note: Click image to view full size file.
Copy number change of PRB4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRB4 | 5545 | 6 | 2 | 22 | |
| GSE20123 | PRB4 | 5545 | 6 | 2 | 22 | |
| GSE43470 | PRB4 | 5545 | 9 | 3 | 31 | |
| GSE46452 | PRB4 | 5545 | 10 | 1 | 48 | |
| GSE47630 | PRB4 | 5545 | 13 | 2 | 25 | |
| GSE54993 | PRB4 | 5545 | 1 | 9 | 60 | |
| GSE54994 | PRB4 | 5545 | 10 | 2 | 41 | |
| GSE60625 | PRB4 | 5545 | 0 | 1 | 10 | |
| GSE74703 | PRB4 | 5545 | 9 | 2 | 25 | |
| GSE74704 | PRB4 | 5545 | 4 | 1 | 15 | |
| TCGA | PRB4 | 5545 | 39 | 7 | 50 |
Total number of gains: 107; Total number of losses: 32; Total Number of normals: 349.
Somatic mutations of PRB4:
Generating mutation plots.
Highly correlated genes for PRB4:
Showing top 20/985 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRB4 | PRH2 | 0.947207 | 3 | 0 | 3 |
| PRB4 | PRB1 | 0.752781 | 12 | 0 | 11 |
| PRB4 | GSX1 | 0.734121 | 4 | 0 | 4 |
| PRB4 | HNF1B | 0.712268 | 8 | 0 | 8 |
| PRB4 | CBLN1 | 0.709326 | 5 | 0 | 5 |
| PRB4 | LPO | 0.707312 | 9 | 0 | 8 |
| PRB4 | PRB3 | 0.689182 | 10 | 0 | 8 |
| PRB4 | CACNG4 | 0.68659 | 9 | 0 | 8 |
| PRB4 | KIR2DS1 | 0.686493 | 7 | 0 | 7 |
| PRB4 | ART1 | 0.686269 | 5 | 0 | 5 |
| PRB4 | SIGLEC7 | 0.682126 | 6 | 0 | 6 |
| PRB4 | ARL5C | 0.681618 | 3 | 0 | 3 |
| PRB4 | NOX5 | 0.681301 | 5 | 0 | 4 |
| PRB4 | MAT1A | 0.678821 | 4 | 0 | 4 |
| PRB4 | FAM223B | 0.678233 | 3 | 0 | 3 |
| PRB4 | PHGDH | 0.678121 | 3 | 0 | 3 |
| PRB4 | PNPLA2 | 0.677549 | 9 | 0 | 7 |
| PRB4 | THEG | 0.677152 | 8 | 0 | 8 |
| PRB4 | VWA3A | 0.675367 | 5 | 0 | 5 |
| PRB4 | NLRP8 | 0.67486 | 3 | 0 | 3 |
For details and further investigation, click here