| Full name: NK6 homeobox 3 | Alias Symbol: FLJ25169 | ||
| Type: protein-coding gene | Cytoband: 8p11.21 | ||
| Entrez ID: 157848 | HGNC ID: HGNC:26328 | Ensembl Gene: ENSG00000165066 | OMIM ID: 610772 |
| Drug and gene relationship at DGIdb | |||
Expression of NKX6-3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NKX6-3 | 157848 | 1552885_a_at | -0.1403 | 0.5251 | |
| GSE26886 | NKX6-3 | 157848 | 1552885_a_at | -0.1104 | 0.4543 | |
| GSE45670 | NKX6-3 | 157848 | 1552885_a_at | 0.1008 | 0.2441 | |
| GSE53622 | NKX6-3 | 157848 | 105998 | 0.3067 | 0.0000 | |
| GSE53624 | NKX6-3 | 157848 | 105998 | 0.2178 | 0.0016 | |
| GSE63941 | NKX6-3 | 157848 | 1552885_a_at | 0.0930 | 0.4728 | |
| GSE77861 | NKX6-3 | 157848 | 1552885_a_at | -0.2105 | 0.0896 | |
| GSE97050 | NKX6-3 | 157848 | A_33_P3421275 | -0.0642 | 0.7435 |
Upregulated datasets: 0; Downregulated datasets: 0.
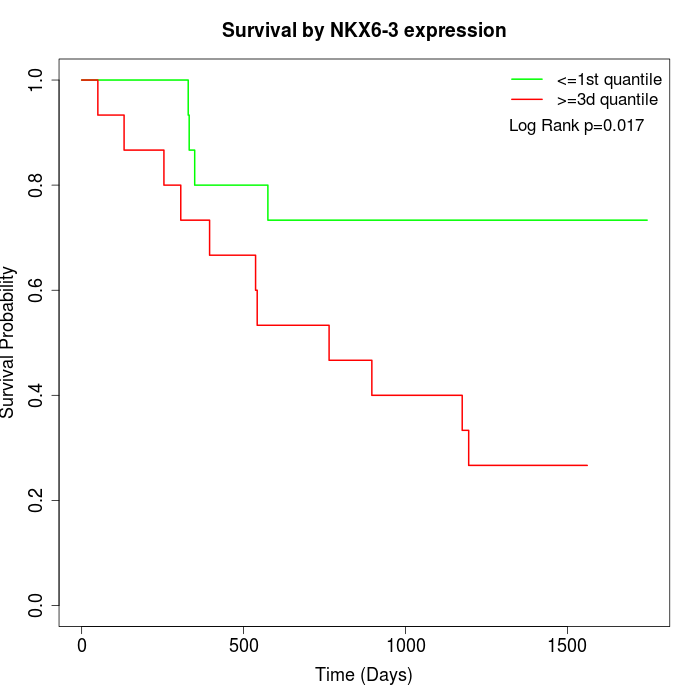
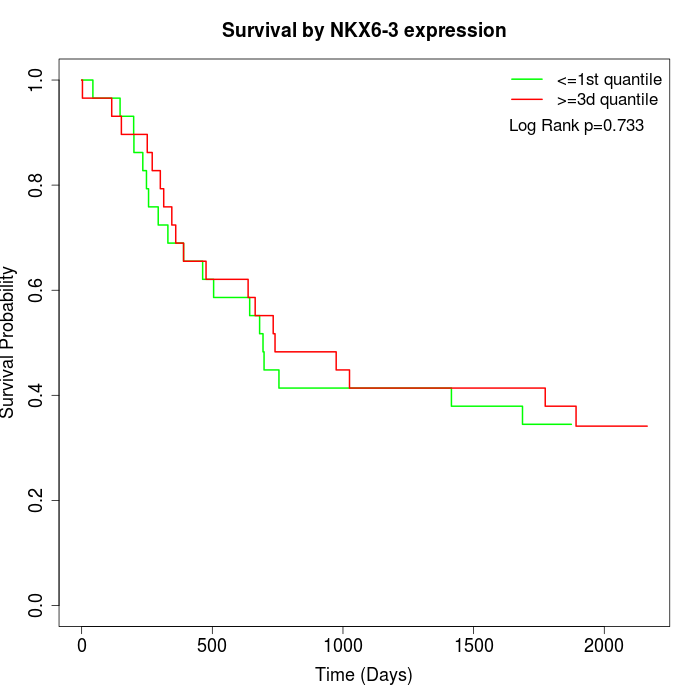
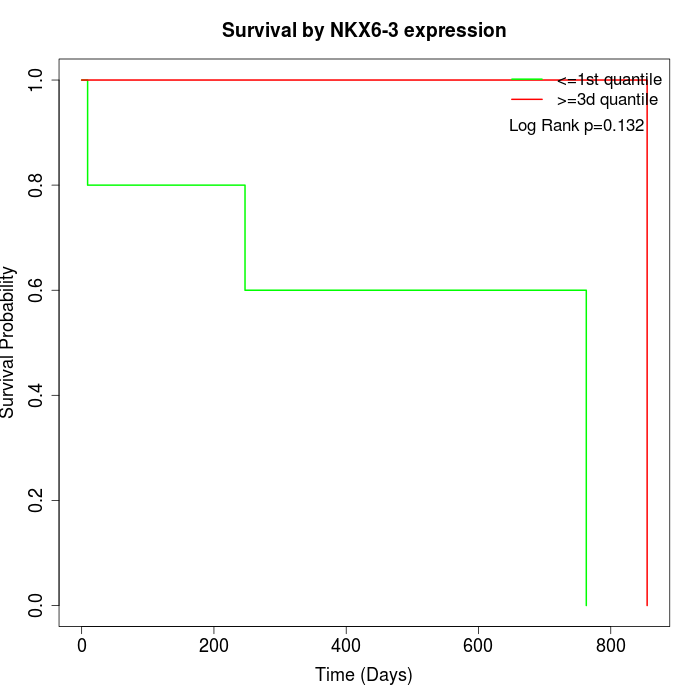
Survival by NKX6-3 expression:
Note: Click image to view full size file.
Copy number change of NKX6-3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NKX6-3 | 157848 | 10 | 5 | 15 | |
| GSE20123 | NKX6-3 | 157848 | 10 | 6 | 14 | |
| GSE43470 | NKX6-3 | 157848 | 9 | 6 | 28 | |
| GSE46452 | NKX6-3 | 157848 | 19 | 5 | 35 | |
| GSE47630 | NKX6-3 | 157848 | 22 | 1 | 17 | |
| GSE54993 | NKX6-3 | 157848 | 1 | 16 | 53 | |
| GSE54994 | NKX6-3 | 157848 | 18 | 8 | 27 | |
| GSE60625 | NKX6-3 | 157848 | 3 | 0 | 8 | |
| GSE74703 | NKX6-3 | 157848 | 8 | 5 | 23 | |
| GSE74704 | NKX6-3 | 157848 | 9 | 2 | 9 |
Total number of gains: 109; Total number of losses: 54; Total Number of normals: 229.
Somatic mutations of NKX6-3:
Generating mutation plots.
Highly correlated genes for NKX6-3:
Showing top 20/455 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NKX6-3 | DCDC1 | 0.904087 | 3 | 0 | 3 |
| NKX6-3 | MRGPRG | 0.883914 | 3 | 0 | 3 |
| NKX6-3 | STRC | 0.87199 | 3 | 0 | 3 |
| NKX6-3 | ZNF763 | 0.866306 | 3 | 0 | 3 |
| NKX6-3 | WDR87 | 0.858055 | 3 | 0 | 3 |
| NKX6-3 | CIB3 | 0.856911 | 3 | 0 | 3 |
| NKX6-3 | YY2 | 0.855492 | 3 | 0 | 3 |
| NKX6-3 | SPACA3 | 0.845402 | 3 | 0 | 3 |
| NKX6-3 | C1QL2 | 0.845392 | 3 | 0 | 3 |
| NKX6-3 | PLA2G2C | 0.844792 | 3 | 0 | 3 |
| NKX6-3 | C9orf153 | 0.8443 | 3 | 0 | 3 |
| NKX6-3 | C2CD4D | 0.831285 | 3 | 0 | 3 |
| NKX6-3 | BBC3 | 0.823459 | 3 | 0 | 3 |
| NKX6-3 | SPTBN4 | 0.822598 | 3 | 0 | 3 |
| NKX6-3 | LHB | 0.822567 | 3 | 0 | 3 |
| NKX6-3 | C19orf71 | 0.821759 | 3 | 0 | 3 |
| NKX6-3 | OR2T27 | 0.819068 | 3 | 0 | 3 |
| NKX6-3 | TCTE1 | 0.817243 | 3 | 0 | 3 |
| NKX6-3 | TRPM8 | 0.80956 | 3 | 0 | 3 |
| NKX6-3 | ZAN | 0.808862 | 3 | 0 | 3 |
For details and further investigation, click here