| Full name: neutral sphingomyelinase activation associated factor | Alias Symbol: FAN|GRAMD5 | ||
| Type: protein-coding gene | Cytoband: 8q12.1 | ||
| Entrez ID: 8439 | HGNC ID: HGNC:8017 | Ensembl Gene: ENSG00000035681 | OMIM ID: 603043 |
| Drug and gene relationship at DGIdb | |||
NSMAF involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04071 | Sphingolipid signaling pathway |
Expression of NSMAF:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NSMAF | 8439 | 203269_at | 0.6167 | 0.1225 | |
| GSE20347 | NSMAF | 8439 | 203269_at | 0.4104 | 0.0441 | |
| GSE23400 | NSMAF | 8439 | 203269_at | 0.4447 | 0.0000 | |
| GSE26886 | NSMAF | 8439 | 203269_at | -0.4230 | 0.0582 | |
| GSE29001 | NSMAF | 8439 | 203269_at | 0.3947 | 0.0897 | |
| GSE38129 | NSMAF | 8439 | 203269_at | 0.3439 | 0.0107 | |
| GSE45670 | NSMAF | 8439 | 203269_at | 0.0831 | 0.6806 | |
| GSE53622 | NSMAF | 8439 | 61896 | 0.2223 | 0.0011 | |
| GSE53624 | NSMAF | 8439 | 61896 | 0.2735 | 0.0000 | |
| GSE63941 | NSMAF | 8439 | 203269_at | -0.8591 | 0.1365 | |
| GSE77861 | NSMAF | 8439 | 203269_at | 0.4239 | 0.0805 | |
| GSE97050 | NSMAF | 8439 | A_23_P134809 | -0.3688 | 0.1682 | |
| SRP007169 | NSMAF | 8439 | RNAseq | 1.1253 | 0.0026 | |
| SRP008496 | NSMAF | 8439 | RNAseq | 0.9732 | 0.0002 | |
| SRP064894 | NSMAF | 8439 | RNAseq | 0.4117 | 0.0091 | |
| SRP133303 | NSMAF | 8439 | RNAseq | 0.2624 | 0.0315 | |
| SRP159526 | NSMAF | 8439 | RNAseq | 0.1673 | 0.5838 | |
| SRP193095 | NSMAF | 8439 | RNAseq | 0.3104 | 0.0004 | |
| SRP219564 | NSMAF | 8439 | RNAseq | 0.0607 | 0.7822 | |
| TCGA | NSMAF | 8439 | RNAseq | 0.0855 | 0.1116 |
Upregulated datasets: 1; Downregulated datasets: 0.
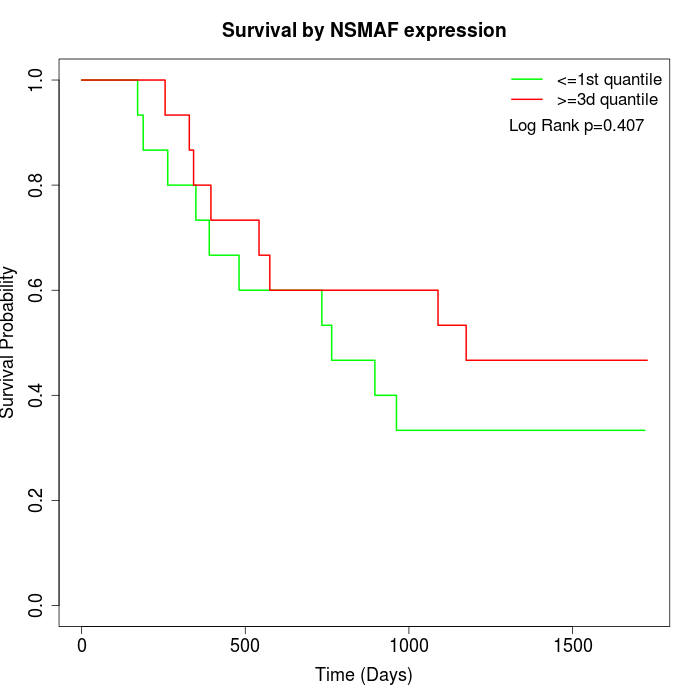
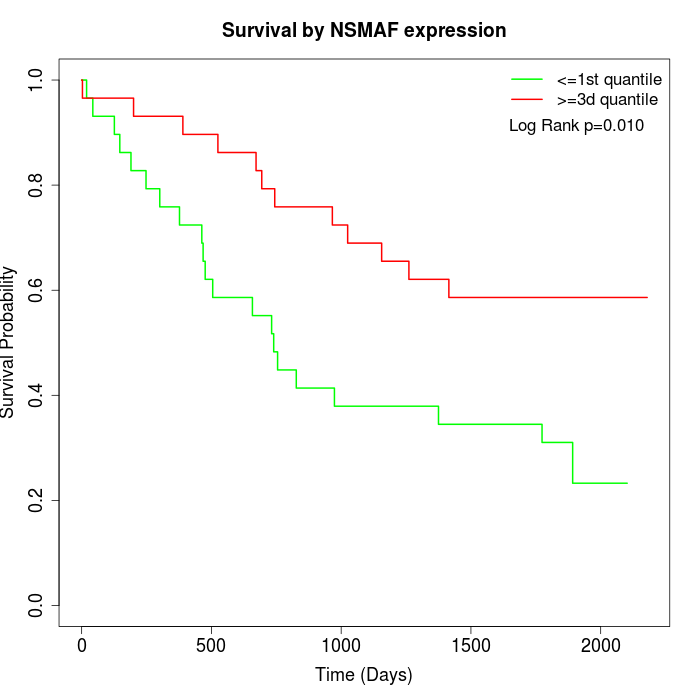
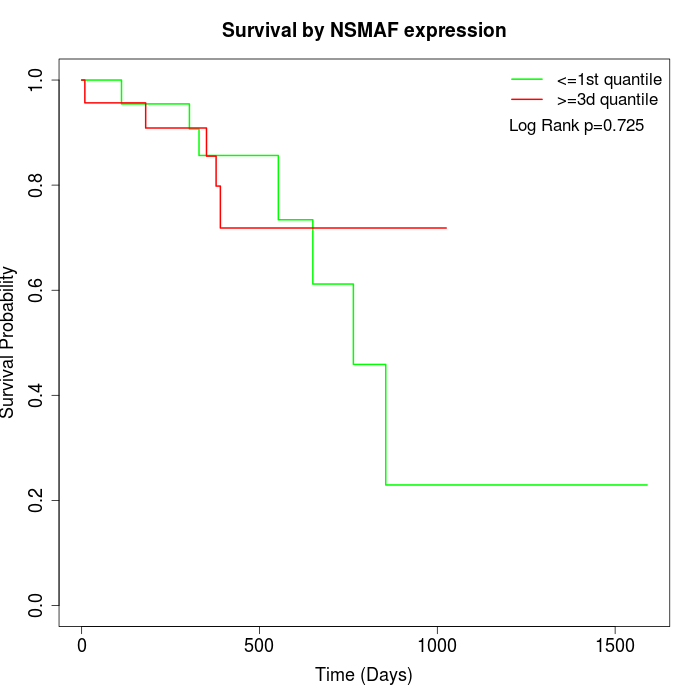
Survival by NSMAF expression:
Note: Click image to view full size file.
Copy number change of NSMAF:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NSMAF | 8439 | 13 | 1 | 16 | |
| GSE20123 | NSMAF | 8439 | 13 | 1 | 16 | |
| GSE43470 | NSMAF | 8439 | 14 | 3 | 26 | |
| GSE46452 | NSMAF | 8439 | 20 | 3 | 36 | |
| GSE47630 | NSMAF | 8439 | 23 | 1 | 16 | |
| GSE54993 | NSMAF | 8439 | 1 | 17 | 52 | |
| GSE54994 | NSMAF | 8439 | 30 | 0 | 23 | |
| GSE60625 | NSMAF | 8439 | 0 | 4 | 7 | |
| GSE74703 | NSMAF | 8439 | 12 | 2 | 22 | |
| GSE74704 | NSMAF | 8439 | 9 | 0 | 11 | |
| TCGA | NSMAF | 8439 | 44 | 6 | 46 |
Total number of gains: 179; Total number of losses: 38; Total Number of normals: 271.
Somatic mutations of NSMAF:
Generating mutation plots.
Highly correlated genes for NSMAF:
Showing top 20/498 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NSMAF | SRRD | 0.839444 | 3 | 0 | 3 |
| NSMAF | WDR75 | 0.793591 | 3 | 0 | 3 |
| NSMAF | MRPS5 | 0.791359 | 3 | 0 | 3 |
| NSMAF | ZNF343 | 0.76156 | 4 | 0 | 4 |
| NSMAF | RLIM | 0.760507 | 3 | 0 | 3 |
| NSMAF | ZNF354A | 0.748965 | 3 | 0 | 3 |
| NSMAF | SLC7A6OS | 0.747303 | 3 | 0 | 3 |
| NSMAF | SNX13 | 0.73379 | 3 | 0 | 3 |
| NSMAF | ZDHHC7 | 0.731476 | 3 | 0 | 3 |
| NSMAF | SRPK2 | 0.715011 | 3 | 0 | 3 |
| NSMAF | TRIM35 | 0.71037 | 4 | 0 | 3 |
| NSMAF | NIPBL | 0.706661 | 3 | 0 | 3 |
| NSMAF | PET117 | 0.703968 | 4 | 0 | 3 |
| NSMAF | MED29 | 0.702861 | 3 | 0 | 3 |
| NSMAF | B4GALT2 | 0.701803 | 3 | 0 | 3 |
| NSMAF | HARS2 | 0.701191 | 3 | 0 | 3 |
| NSMAF | RFWD3 | 0.69984 | 3 | 0 | 3 |
| NSMAF | CLK2 | 0.699783 | 4 | 0 | 4 |
| NSMAF | UCKL1 | 0.698979 | 3 | 0 | 3 |
| NSMAF | ZNF451 | 0.69866 | 3 | 0 | 3 |
For details and further investigation, click here