| Full name: NIPBL cohesin loading factor | Alias Symbol: IDN3|DKFZp434L1319|FLJ11203|FLJ12597|FLJ13354|FLJ13648|Scc2 | ||
| Type: protein-coding gene | Cytoband: 5p13.2 | ||
| Entrez ID: 25836 | HGNC ID: HGNC:28862 | Ensembl Gene: ENSG00000164190 | OMIM ID: 608667 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of NIPBL:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NIPBL | 25836 | 213918_s_at | 0.4113 | 0.4019 | |
| GSE20347 | NIPBL | 25836 | 213918_s_at | -0.1239 | 0.5135 | |
| GSE23400 | NIPBL | 25836 | 212469_at | 0.1194 | 0.0050 | |
| GSE26886 | NIPBL | 25836 | 213918_s_at | 0.5538 | 0.0337 | |
| GSE29001 | NIPBL | 25836 | 213918_s_at | -0.0223 | 0.9434 | |
| GSE38129 | NIPBL | 25836 | 213918_s_at | 0.1540 | 0.4365 | |
| GSE45670 | NIPBL | 25836 | 213918_s_at | 0.2559 | 0.1735 | |
| GSE53622 | NIPBL | 25836 | 15822 | 0.0977 | 0.1473 | |
| GSE53624 | NIPBL | 25836 | 15822 | 0.0206 | 0.7395 | |
| GSE63941 | NIPBL | 25836 | 213918_s_at | 0.3652 | 0.4540 | |
| GSE77861 | NIPBL | 25836 | 213918_s_at | 0.1700 | 0.3989 | |
| GSE97050 | NIPBL | 25836 | A_23_P213883 | 0.0200 | 0.9272 | |
| SRP007169 | NIPBL | 25836 | RNAseq | 0.1859 | 0.5628 | |
| SRP008496 | NIPBL | 25836 | RNAseq | 0.0123 | 0.9544 | |
| SRP064894 | NIPBL | 25836 | RNAseq | -0.3721 | 0.0674 | |
| SRP133303 | NIPBL | 25836 | RNAseq | 0.0532 | 0.6494 | |
| SRP159526 | NIPBL | 25836 | RNAseq | 0.3425 | 0.3379 | |
| SRP193095 | NIPBL | 25836 | RNAseq | -0.2338 | 0.0186 | |
| SRP219564 | NIPBL | 25836 | RNAseq | -0.3988 | 0.1718 | |
| TCGA | NIPBL | 25836 | RNAseq | 0.0260 | 0.5921 |
Upregulated datasets: 0; Downregulated datasets: 0.
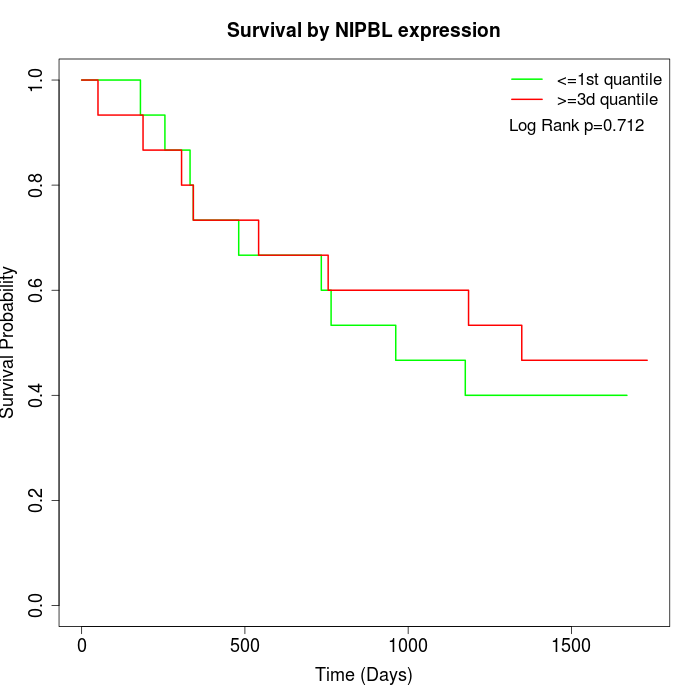
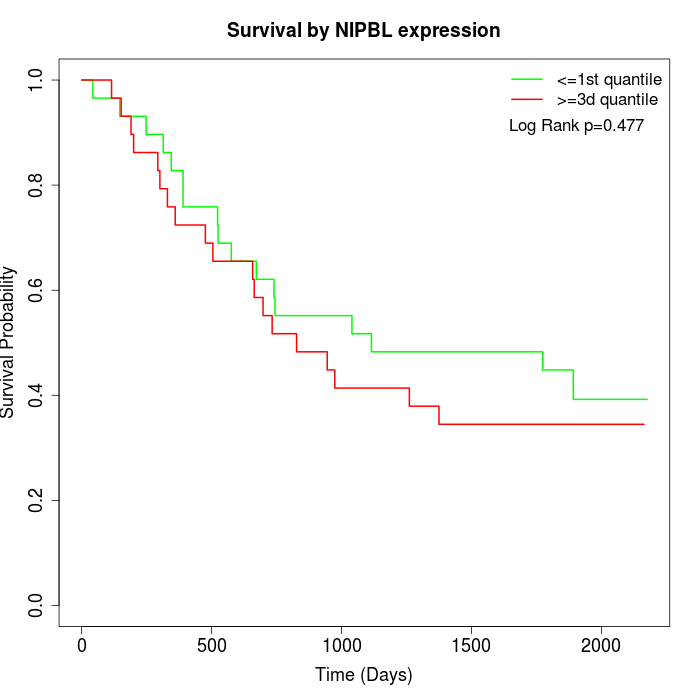
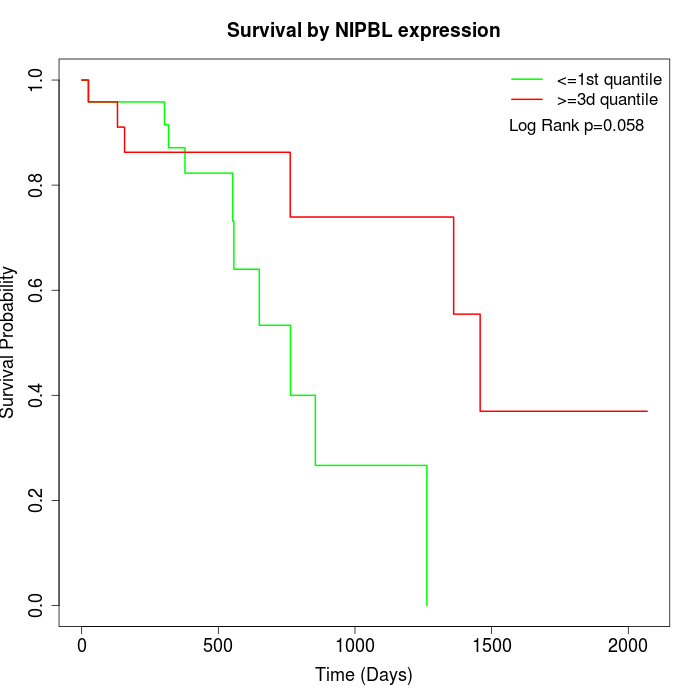
Survival by NIPBL expression:
Note: Click image to view full size file.
Copy number change of NIPBL:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NIPBL | 25836 | 10 | 0 | 20 | |
| GSE20123 | NIPBL | 25836 | 10 | 0 | 20 | |
| GSE43470 | NIPBL | 25836 | 18 | 0 | 25 | |
| GSE46452 | NIPBL | 25836 | 5 | 22 | 32 | |
| GSE47630 | NIPBL | 25836 | 6 | 12 | 22 | |
| GSE54993 | NIPBL | 25836 | 5 | 4 | 61 | |
| GSE54994 | NIPBL | 25836 | 26 | 2 | 25 | |
| GSE60625 | NIPBL | 25836 | 0 | 0 | 11 | |
| GSE74703 | NIPBL | 25836 | 14 | 0 | 22 | |
| GSE74704 | NIPBL | 25836 | 9 | 0 | 11 | |
| TCGA | NIPBL | 25836 | 53 | 6 | 37 |
Total number of gains: 156; Total number of losses: 46; Total Number of normals: 286.
Somatic mutations of NIPBL:
Generating mutation plots.
Highly correlated genes for NIPBL:
Showing top 20/241 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NIPBL | TRUB2 | 0.762862 | 3 | 0 | 3 |
| NIPBL | EPRS | 0.756074 | 3 | 0 | 3 |
| NIPBL | AP1G1 | 0.755683 | 3 | 0 | 3 |
| NIPBL | KIAA0895L | 0.751498 | 3 | 0 | 3 |
| NIPBL | C11orf80 | 0.749798 | 3 | 0 | 3 |
| NIPBL | ZNF700 | 0.739455 | 3 | 0 | 3 |
| NIPBL | OSBPL2 | 0.736314 | 4 | 0 | 4 |
| NIPBL | HSPA4 | 0.732957 | 3 | 0 | 3 |
| NIPBL | RAB13 | 0.73265 | 3 | 0 | 3 |
| NIPBL | NUDT19 | 0.727958 | 3 | 0 | 3 |
| NIPBL | KDM2A | 0.727323 | 3 | 0 | 3 |
| NIPBL | FAM49B | 0.724542 | 3 | 0 | 3 |
| NIPBL | WIPI2 | 0.723088 | 3 | 0 | 3 |
| NIPBL | OTUD6B | 0.718095 | 3 | 0 | 3 |
| NIPBL | MRPS18B | 0.715513 | 3 | 0 | 3 |
| NIPBL | NIP7 | 0.715107 | 3 | 0 | 3 |
| NIPBL | NAA15 | 0.714086 | 3 | 0 | 3 |
| NIPBL | MZT2B | 0.71377 | 3 | 0 | 3 |
| NIPBL | EXOSC10 | 0.70938 | 3 | 0 | 3 |
| NIPBL | ZNF121 | 0.707597 | 4 | 0 | 3 |
For details and further investigation, click here